Detection and Characterization of Extractables in Food Packaging Materials by GC–MS
Special Issues
In this study, general extract screening of food storage materials was done with nontargeted analytical methods to understand what analytes could potentially leach into food or beverages. GC and mass spectral deconvolution effectively separated analytes within the complex mixture and TOF-MS provided full mass range spectral data for identification. This workflow can be used for confident characterization of components present as extractables from food packaging materials.
Migration or leaching of analytes from packaging material is a concern for manufacturers and consumers because of the potential contamination of food and beverages. This leaching can impact the quality of the product, affect the integrity of the packaging material, and cause concern related to consumer health and product safety. To investigate analytes with the potential for leaching, an extraction of a variety of food packaging or storage products, including sealable plastic bags and plastic food containers, was performed. Solvent was placed inside each packaging product for an extended period of time and then concentrated through evaporation before analysis. General screening of this extract with nontargeted analytical techniques was used to understand what analytes were extracted and may have the potential to leach into the food. Gas chromatography (GC) was used for separating analytes from each other and time-of-flight mass spectrometry (TOF-MS) provided full mass range spectral data. Nominal-mass TOF-MS data were acquired and searched against library databases for tentative identifications. High-resolution TOF-MS data were also acquired to add confidence to identifications with accurate mass information. Several analytes were determined and are highlighted here.
Polymer-based materials are often used to manufacture food packaging and disposable food storage containers. These types of materials tend to be lightweight, flexible, and durable, which makes them desirable for this purpose and for many other uses. To add to their versatility and achieve the required characteristics, additives are typically incorporated into the polymer. These can be processing aids, long-term thermal stabilizers, or ultraviolet (UV) protectors that are all intended to protect the polymers and prevent degradation both during production and use. Other modifiers that alter or improve the properties for use, such as plasticizers, lubricants, or coloring agents, are also common (1).
These additives are sometimes pure, but can be mixed with other additives and base materials or derived from sources with potentially unknown by-products. Any of these unintended analytes can also become incorporated into the polymer. Degradation or transformation of additives is also known to occur and can be expected in the finished product (1). For these reasons, polymer-based materials can become fairly complex samples that contain a wide range of molecules that have the potential to leach from the packaging or storage container into the food or beverage product. This leaching can lead to the inadvertent consumption by a consumer, which may be a health and safety concern (2).
Studies that intentionally extract polymer-based materials with solvent and harsh storage conditions can provide insight to what molecules have the potential to leach into food or beverage products. These studies represent worst-case scenarios and are important for understanding what is in a product. In this work, we used gas chromatography (GC) to separate individual extracted analytes and paired it with time-of-flight mass spectrometry (TOF-MS) and high-resolution TOF-MS to identify the extracted and separated analytes.
Experimental
Sample Preparation
Analytes were extracted from a variety of food storage products, including three brands of flexible bags and one plastic reusable box. First, 100 mL of a 1:1 hexane–acetone solution was placed inside each food storage container for 20 h at room temperature to obtain potential extractables. The solvent was then evaporated to 1 mL under nitrogen before analysis.
Instrumental Analysis
The concentrated solvent was then analyzed by GC–TOF-MS with electron ionization (EI) (Pegasus BT, LECO) and by GC–HR-TOFMS with EI and chemical ionization (CI) (Pegasus HRT, LECO). Instrument conditions are listed in Tables I and II.
Data Analysis
LECO’s ChromaTOF brand software was used for peak finding, deconvolution, library searching, retention index calculations, and formula determinations for accurate-mass data.
Results and Discussion
A variety of storage materials were analyzed to determine extractable analytes that have the potential to leach into food products. Each sample was analyzed by GC paired with a nominal-mass TOF-MS system with EI, a high-resolution TOF-MS system with EI, and a high-resolution TOF-MS system with CI. This data collection strategy provided chromatographic separations of individual analytes and their mass spectra for both library searching and formula determinations. Analysis across instrument platforms was valuable for determining analytes and their identifications and also provided the opportunity to explore benefits of accurate mass data for these types of samples. Representative nominal mass chromatograms for each sample are shown in Figure 1.
Figure 1: Representative TIC chromatograms from the GC–TOF-MS data are shown for each food storage product. Three flexible bags (Brand A, B, and C) and a reusable plastic box (Brand A) are shown.
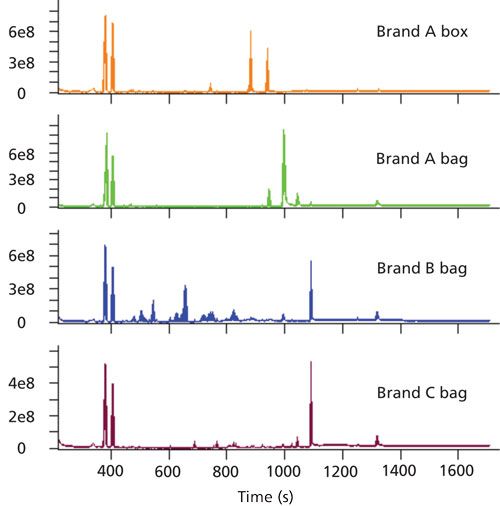
Hundreds of analytes were separated from each other and detected with this analytical approach. The separation was achieved through a combination of chromatography and deconvolution, which takes advantage of the full mass range data to add mathematical separation in instances of chromatographic coelution. For complex samples, this approach is beneficial because chromatographic coelution is common. An example of deconvolution is demonstrated in Figure 2 where a plasticizer and slip agent chromatographically are coeluted, but are mathematically separated. In Figure 2, the total ion current chromatogram (TIC) peak (scaled to 2% for visualization) and extracted-ion chromatogram (XIC) traces (m/z 277.18 and 256.24) that correspond to each analyte are overlaid. It is not evident from the TIC that more than one analyte is eluted at this retention time, but the two analytes can be observed in the XIC traces. These analytes have a large concentration difference, yet automated deconvolution still determined that both were present and provided clean spectral information for each. The mass spectral data allowed for determining the identifications of each analyte and the identification workflow is highlighted in Figure 2.
Figure 2: Deconvolution separates Metilox and palmitic acid that are chromatographically coeluted. The identification workflow of these analytes combined library searching of nominal-mass spectra and retention index verification from TOF-MS data, and accurate-mass formula determinations from both EI and CI HR-TOF-MS data.

Library searching of nominal-mass data, retention index determination with comparison to database values, and formula determinations from EI and CI accurate-mass data were combined to determine analyte identifications. Library searching of the nominal-mass spectral data provided the tentative identification of Metilox, a plasticizer, with a similarity score of 827 compared to the National Institute of Standards and Technology (NIST) libraries. This identification was supported by the retention index with 0.98% deviation between the calculated value, 1962, and the NIST (semistandard nonpolar) reference value, 1943 (3). To add further confidence to these identifications, accurate-mass data were acquired with a high-resolution TOF-MS system. The EI and CI spectra for this analyte are also shown. The EI spectrum was library searched against the NIST database with a similarity score of 809 and formulas were determined for the molecular ion and several fragments. The observed and theoretical values are compared in the associated table with excellent mass accuracies. The molecular ion, C18H28O3, was also observed with a mass accuracy of 1.22 ppm. The CI data further supported this molecular ion and also included the common methane adduct (M+C2H5) with a mass accuracy of 0.57 ppm. In this case, the accurate-mass data supported the nominal-mass identification and all of the information combined to give greater confidence in the preliminary identification of this plasticizer. That is also the case for the coeluted slip agent, shown in Figure 2. Library searching provided palmitic acid as the tentative identification with a similarity score of 936 compared to the NIST libraries. The retention index supported this identification with a 0.05% deviation between the calculated value of 1969 and the NIST value of 1968. The EI and CI spectra are also shown. The EI had a similarity score of 928 compared to the NIST libraries. The molecular ion, C16H32O2, and numerous fragments were observed with excellent mass accuracy. The CI data supported the molecular ion determination with both the M+H and a common methane adduct (M+C2H5) observed.
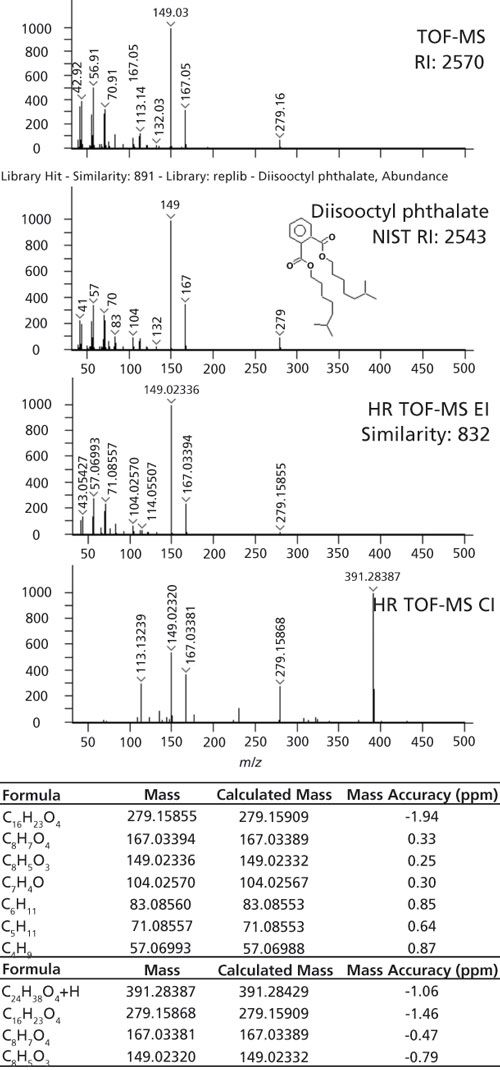
When adding analytical capabilities, like high-resolution TOF-MS with EI and supplemental CI, there is the potential to improve identifications as well as overall confidence in those identifications. In Figure 3, an example is highlighted where CI data helped clarify the identification by adding molecular ion information that was not observed with EI. Library searching yielded a tentative identification of diisooctyl phthalate with a similarity score of 891 compared to NIST library spectra. This identification was supported by retention index as the calculated value of 2570 had 1.06% deviation compared to the NIST value of 2543. The accurate mass EI data also matched to diisooctyl phthalate with a similarity score of 832. Several fragments with excellent mass accuracy were verified with formula determinations, but the molecular ion, C24H38O4, was not observed. CI data provided the M+H ion, with good mass accuracy to give greater confidence in the preliminary identification of this phthalate.
Figure 3: The identification workflow is demonstrated for diisooctyl phthalate. Library searching, retention index verification, and formula determinations assisted in the identification. CI added information on the molecular ion that was not observed with EI.
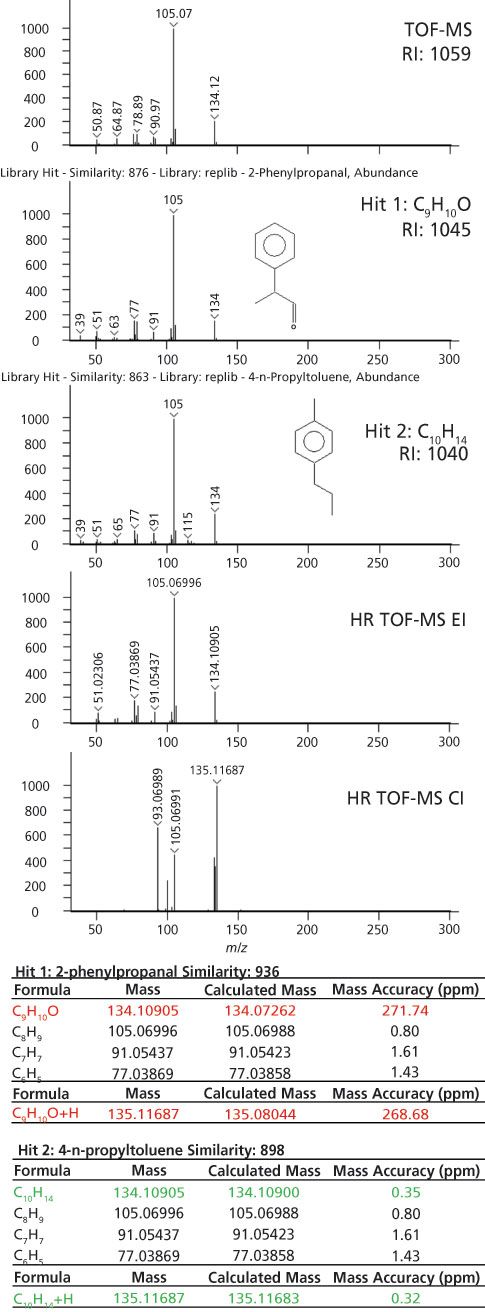
In other instances, the accurate-mass information was crucial for sorting out uncertainty in the nominal-mass identifications. An example is highlighted in Figure 4 where two library matches were reasonably based on the library search and retention index results. The nominal-mass spectrum is shown in the top part of Figure 4. This analyte had a calculated retention index of 1059. The first library hit had a similarity score of 876 with a retention index value of 1045 (1.3% error) and the second library hit had a similarity score of 863 with a retention index value of 1040 (1.8% error). Both of these identifications have the same nominal molecular weight (134 m/z) and many of the same fragments in their EI spectra. The exact masses for these potential identifications, C9H10O and C10H14, are 134.07262 and 134.10900, respectively. This difference of 0.03 Da can be distinguished with high-resolution systems. The high-resolution TOF-MS data had the same first library hit as the nominal mass data, but the calculated formula for the observed mass, 134.10905, had a mass accuracy of 272 ppm when compared to C9H10O. A much better formula to explain this mass was C10H14 with a mass accuracy of 0.35 ppm. The high-resolution TOF-MS data clarified the correct formula and suggested that the correct identification was hit 2. In this instance, where the molecular weight and many fragments had the same nominal mass and the retention index values were similar to each other, the accurate mass information was crucial to determine the correct identification.
Figure 4: The workflow for identification is demonstrated for an analyte that could not be discerned without accurate-mass data. The nominal-mass molecular weight and fragments were the same and the retention index information was very similar. The accurate mass measurements strongly support hit 2 as the correct assignment.
This workflow was used to compile Table III, which contains 33 representative analytes observed in these samples. This table compiles data across the instrument platforms and includes NIST database information. For each analyte, similarity scores compared to NIST library spectra are presented. The observed retention index values were calculated based on alkane elution and compared to the semistandard, nonpolar NIST database values with the percent deviation between the two reported. Molecular ion formulas and mass accuracy are also presented from the high resolution EI data. In the absence of an EI molecular ion, the CI molecular ions are presented as indicated by the formula. The presence or absence of an analyte in each sample is indicated with “x” and the purpose of the analyte as an additive is indicated, when known. Several additives and other analytes of potential interest were extracted and observed in these data. This list is not comprehensive, but it serves as a set of examples of the types of analytes that were extracted with this protocol and observed with these analytical methods.

CLICK TABLE TO ENLARGE
One particular additive pair is highlighted in Figures 5 and 6. Irgafos 168 is an organic phosphite that is an antioxidant and process stabilizer. A known transformation product, Tris(2,4-di-tert-butylphenyl) phosphate, is also shown. Both were observed in these samples and the spectral identification information is shown in Figure 6. The nominal MS data had a similarity score of 863 for Irgafos 168. The EI high-resolution MS data had a similarity score of 899. The molecular ion (observed with both EI and CI) and several fragments are listed with excellent mass accuracy. The same is true for the transformation product. The nominal MS data had a similarity score of 820 and the high-resolution MS similarity score was 791. Mass accuracies for the molecular ion (observed with both EI and CI) and several fragments are listed with excellent mass accuracy. The relative ratios of these two analytes are also shown in Figure 7. Because these analytes are processing stabilizers, the transformation between these two additives and their relative ratios may be of interest to engineers monitoring process controls.
Figure 5: Structures of Irgafos 168 and its transformation product, tris(2,4-di-tert-butylphenyl) phosphate.
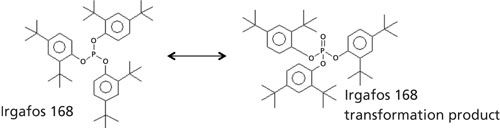
Figure 6: MS analysis of Irgafos 168 and its transformation product, tris(2,4-di-tert-butylphenyl) phosphate.
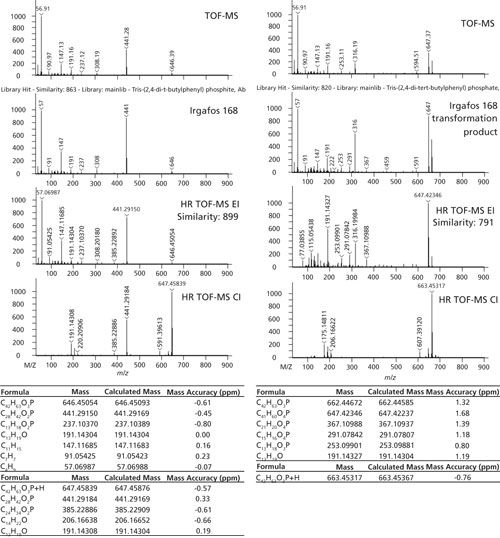
Figure 7: Relative ratio of Irgafos 168 and its associated transformation product in each sample.
Conclusion
This study investigated extracts of food storage materials to gain insight into potential leachable analytes. General extract screening was done with nontargeted analytical methods to understand what analytes could potentially leach into food or beverage products. GC and mass spectral deconvolution effectively separated analytes within the complex mixture and TOF-MS provided full mass range spectral data for identification. Nominal-mass TOF-MS data were searched against library databases for preliminary identifications, which were then confirmed or updated based on retention index information. High-resolution TOF-MS data were then used to further confirm or update identifications with accurate mass information and formula determinations. Several analytes were determined and highlighted. This study demonstrated a workflow that was effectively used to confidently characterize components present as extractables from food packaging materials.
References
- M.A. Ruberto, “Polymers and Additives Used in Fabrication of Disposable Bioprocess Equipment,” Bioprocess International, April 2010 supplement.
- J.D. Meeker, S. Sathyanarayana, and S.H. Swan, Philos. Trans. R. Soc.364(1526), 2097–2113 (2009).
- NIST Mass Spectral Search Program for the NIST/EPA/NIH Mass Spectral Library Version 2.2, build Jun 10 2014.
Elizabeth M. Humston-Fulmer is an applications chemist with LECO Corporation in St. Joseph, Michigan. Joe Binkley is the director of applications lab with LECO Corporation. Direct correspondence to: liz_humston-fulmer@leco.com

Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Revealing the Ancient Secrets of Chinese Swamp Cypress Using Cutting-Edge Pyrolysis Technology
November 18th 2024A study published in the Journal of Analytical and Applied Pyrolysis by Yuanwen Kuang and colleagues used advanced pyrolysis techniques to reveal the preservation and chemical transformations of 2,000-year-old Chinese swamp cypress wood, offering valuable insights for archaeological conservation and environmental reconstructions.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.