Development of a High Sensitivity Method for the Analysis of Clopidogrel and Clopidogrel Carboxylic Acid Metabolite in Human K2EDTA Plasma Using UHPLC–MS-MS
The method development process required for the accurate quantification of both clopidogrel and its acid metabolite with a lower limit of quantification (LLOQ) of 1 pg/mL in human plasma is discussed.
Bioanalytical methods often involve the quantification of a parent compound to determine the pharmacokinetic properties of a potential drug. In some cases, it may also be necessary to quantify a metabolism product in addition to the parent molecule as these may be either active or toxic. Simultaneous determination can be challenging because of differences in chemical properties, such as acidity, basicity, and polarity, which can significantly increase the difficulty in developing both the extraction method as well as suitable chromatography. Developing chromatographic conditions involves not only separating the parent from the metabolite, but also effectively separating both compounds from potential coeluted contaminants such as phospholipids that may affect ionization efficiency and cause suppression or enhancement. The stabilities of the parent and metabolite also must be considered, as any interconversion between the two during the bioanalytical procedure may lead to variability within the results. In this article, we discuss the method development process required for the accurate quantification of both clopidogrel and its acid metabolite with a lower limit of quantification (LLOQ) of 1 pg/mL in human plasma.
It is of great importance to integrate pharmacokinetic, pharmacodynamic, and toxicokinetic information in the process of drug development. Serious decisions are often made based on bioanalytical data, so it is vitally important that these data are precise and accurate. Before any bioanalytical method can be used for the analysis of samples acquired from dosed subjects, it is necessary to validate the method according to the appropriate guidelines, set forth by the US Food and Drug Administration (FDA), the European Medicines Agency (EMA), and others. Because of the strict requirements for method validation, it is essential to be thorough during the method development process and to identify any potential issues before the validation process. The best strategy for method development is to take a systematic approach with the three main aspects as follows: the extraction method, the liquid chromatography (LC) method, and the detection method. For the development of a sensitive, robust, and accurate method, all three portions are equally important.
As stated above, many bioanalytical methods require the quantification of more than one component. In many cases, the quantification of one or more metabolism products is essential for obtaining accurate data regarding the drug's pharmacokinetic profile. When an assay requires the quantification of multiple components, the development process becomes more difficult than it would be for a single compound. For some assays, the relative concentration requirements of the parent and metabolite may be vastly different because of the expected levels found in vivo. In this case, it may be best to optimize the method for the compound requiring the lower limit of quantification (LLOQ), and then make any necessary modifications to accommodate the other components.
Clopidogrel (trade name Plavix, Figure 1), is a thienopyridine derivative antiplatelet prodrug used in the prevention of artherosclerotic events. It is dosed in an inactive form and requires a hepatic biotransformation to yield the active thiol-metabolite, which binds to cell receptor P2Y12, irreversibly inhibiting the platelet activation process (1). In addition to the active metabolite, an inactive carboxylic acid metabolite is also formed. This acid metabolite accounts for the majority of circulating clopidogrel-related material with very low levels of the active metabolite and unchanged prodrug present (2,3). Because of the reactivity of the thiol metabolite, coupled with the low levels of the unchanged prodrug, most quantitative studies are based on the circulating levels of the inactive metabolite. The ability to detect the low levels of unchanged prodrug will provide more accurate data on the pharmacokinetics of clopidogrel, allowing improved evaluation of the bioavailability of new formulations.
Figure 1: Chemical structures of clopidogrel and clopidogrel carboxylic acid metabolite.
Experimental
The finalized experimental conditions for the extraction procedure and ultrahigh-pressure liquid chromatography coupled with tandem mass spectrometry (UHPLC–MS-MS) are described below.
Solid-Phase Extraction
Plasma samples were extracted by diluting 350 μL of plasma sample, containing 10 μL of internal standard solution at a concentration of 250 pg/mL, with 350 μL of Milli-Q (Millipore Corporation) water. Samples were added to an Oasis hydrophilic-lipophilic balanced (HLB) μElution plate (Waters Corporation) after preconditioning the plate with 200 μL of methanol followed by 200 μL of water. Samples were drawn through under vacuum, and then washed with 200 μL of water and 200 μL of 5% methanol–water. The sample was eluted with 2 × 25 μL of methanol and then diluted with an equal volume of water before injection. Samples were prepared at the following concentrations: 1.00, 2.50, 5.00, 10.0, 25.0, 50.0, 100, 250, and 500 pg/mL.
UHPLC–MS Conditions
The UHPLC–MS system consisted of an Acquity UPLC system (Waters Corporation) coupled to a tandem quadrupole Xevo TQ-S mass spectrometer (Waters Corporation) equipped with a prototype low-flow capillary for use with 1.0-mm i.d. columns. Chromatographic separations were performed on a 50 mm × 1.00 mm, 1.7-μm dp Waters Acquity UPLC BEH C18 column (Waters Corporation) maintained at 45 °C. Mobile-phase A consisted of 0.1% formic acid and mobile-phase B was 100% acetonitrile. The flow rate was 0.140 mL/min. Starting conditions of 10% B were held for 0.5 min, then components were eluted via a linear gradient of 10–90% B over 2.5 min. The MS system was operated in positive electrospray ionization (ESI+ ) mode, and the following multiple reaction monitoring (MRM) transitions were used: 322.1→212.0 for clopidogrel, 326.1→216.1 for d4-clopidogrel, 308.1→198.1 for clopidogrel acid metabolite, and 312.1→202.1 for the d4-clopidogrel acid metabolite. The capillary and cone voltages were set to 0.5 kV and 35 V, respectively. The optimized collision energies for all four components were determined to be 16 V. All data integration and calculations were done using Waters UNIFI Scientific Information System software (Waters Corporation).
Results and Discussion
Most published methods for the analysis of clopidogrel or its carboxylic acid metabolite use a liquid–liquid extraction (LLE) technique, often requiring a double LLE before UHPLC–MS (3,4). These methods are time consuming and tedious, and require large volumes of harmful chemicals such as hexane and diethyl ether. For these reasons, the use of alternative solid-phase microelution technology was investigated. This methodology provides an increase in throughput while decreasing solvent consumption. A typical LLE will consume 2–8 mL of organic solvent per sample, in contrast to microelution solid-phase extraction (SPE) methods, which require less than 0.5 mL of organic solvent per sample. Because of the fact that the carboxylic acid metabolite is present at much higher concentrations in circulating blood, development of an extraction procedure was optimized for the prodrug, clopidogrel. Results from initial method development experiments showed that mixed-mode cation exchange and HLB were the two most promising SPE sorbents to further optimize. Both the analyte and metabolite were shown to extract well using these methods, and there was no loss shown of either component during the wash steps. Use of the microelution plate made it possible to load 350 μL of plasma sample (diluted 1:1 with aqueous solution before loading), and elute the analytes with only 50 μL of solvent. This gives a sevenfold increase in concentration and also eliminates the need for a time-consuming dry down step that is found in most bioanalytical methods, especially LLEs. Because of the chromatographic differences in the two compounds (discussed later in this article), a 1:1 dilution of the SPE eluent with water was required before injection.
The next step in the method development process was to conduct a mobile-phase screen to determine the mobile-phase composition that would provide both the best MS ionization characteristics and separation of clopidogrel from any coeluted interferences. Detection was carried out on a triple-quadrupole mass spectrometer operating in ESI+ conditions. The initial screening was conducted on a 50 mm × 2.1 mm, 1.7-μm dp UHPLC C18 column. The results for all combinations of either acidic or basic aqueous mobile phase modifier when used in combination with acetonitrile or methanol as the organic eluent indicate that 0.1% ammonium hydroxide with acetonitrile gave the greatest response as well as the greatest signal-to-noise value. Although a majority of mobile phases used in LC–MS are traditionally acidic, such as 0.1% formic acid, many recent studies show that for a wide range of pharmaceutical compounds, high pH often provides the best conditions for ionization (5).
Typically, a method developer seeks to find chromatographic conditions that yield the highest signal-to-noise ratios. This provides a lower detection limit, which is often the limiting factor in a bioanalytical method. However, one must also take into account any coeluted interferences that may potentially impact the assay via suppression, by enhancement, or by producing variability within the assay. Some of the most common interferences within a bioanalytical assay arise from the choline-containing lipids found in the matrix, which share a common fragmentation of 184 m/z. Using this piece of information, it is possible to monitor a majority of these compounds either by using a generic 184→184 m/z MRM transition or by collecting precursor data. For clopidogrel, acquisition of both clopidogrel MRM 322→212 m/z along with a precursor scan (precursors of m/z 184, from 450 m/z to 800 m/z) were collected simultaneously. Evaluation of the acquired data for both the acidic and basic aqueous mobile phase combined with acetonitrile showed that clopidogrel is eluted in the middle of the phospholipid region when basic solution is used. In contrast, for 0.1% formic acid with acetonitrile, clopidogrel is on the leading edge of the phospholipid region. In this case, it is much easier to chromatographically separate the clopidogrel from the phospholipid fraction. Although the response of the clopidogrel was not obviously suppressed by the coelution of lipids, it can still lead to inconsistent data because of varying amounts of lipids within the sample, or varying interactions with the lipids that may affect ionization. Both of these scenarios have the potential to change on a sample-by-sample basis, which may lead to assay irreproducibility. For these reasons, the use of ammonium hydroxide with acetonitrile was not used for further method optimization.
Because of the fact that phospholipids are not the only potential interferences, the full scan data as well as the 184 m/z precursor scan data were used to determine the best column choice. The UHPLC 1.7-μm dp C18 column showed the lowest response for both the precursors of 184 m/z and full-scan data in the region of the clopidogrel peak elution. The next step was the development of the LC conditions to accommodate both the clopidogrel and its acid metabolite. In light of the differences in structure and hydrophobicity, the elution composition for each component was drastically different. Because of the hydrophobic property of clopidogrel, it is advantageous to use LC gradient starting conditions with a higher percentage of organic solvent to expedite the elution and thus shorten the run time. In contrast, the acid metabolite is eluted much earlier in the gradient profile, thus if starting conditions contain too much organic, the k (retention factor) value will not be optimal and peak shape may suffer. These factors also had to be taken into account before introduction of the sample into the UHPLC system. If the injection solution percent organic is too high, the peak shape of the acid metabolite may potentially suffer via early elution. For these reasons, the development of the optimal method contained a 90:10 aqueous–organic hold for 0.5 min before the gradient start. This provided a k value of 1.6 for the metabolite and resulted in no discernible band spreading for the 50:50 aqueous–organic injection.
By using a microelution SPE plate for sample preparation, the resulting bioanalytical method yielded an LLOQ of 1 pg/mL for both the parent and acid metabolite. The final method used d4-labeled internal standards for both clopidogrel and its carboxylic acid metabolite to account for any variability within the extraction and detection. The next step in creating a bioanalytical method that is appropriate for use is to test for robustness and reproducibility. In a regulated setting where the method would be used to analyze study samples, a formal method validation would be required before use. After several accuracy and precision batches were extracted and injected, it was noticed that the results for both the parent and metabolite were very erratic, especially near the lower end of the calibration range. To determine whether it was an effect of some components in the plasma, the extraction was performed using calibration standards and quality controls (QCs) prepared both in human plasma and in aqueous solution. The results showed the same nonreproducibility for both matrices, indicating that the unreliable results were not because of any of the matrix components. Replicate injections of the same sample, whether in plasma or in solution, produced MS responses with very low variability. This indicated that the erratic results were not because of the UHPLC–MS portion of the analysis. For example, any problem with column re-equilibration, insufficient ionization, or hardware (such as leaks) would have been noticed when injecting replicates of the same sample. The problem, therefore, seemed to lie somewhere within the extraction method; thus an alternative HLB reversed-phase extraction method was investigated.
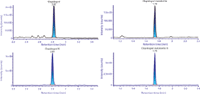
Figure 2: Example chromatograms of clopidogrel (top left), deuterated clopidogrel (bottom left), carboxylic acid metabolite (top right), and deuterated carboxylic acid metabolite (bottom right).
From initial method development, the use of the HLB sorbent gave the second best recovery and signal to noise values. In addition, the use of HLB sorbent did not produce acidic or basic extracts, which may have been the cause of the nonreproducibility when using the mixed mode cation exchange sorbent plate. Because it is the methyl ester of clopidogrel that undergoes a metabolism to yield the carboxylic acid metabolite, it is reasonable to assume that there may be conversion or back-conversion within the extraction procedure. Even if the reaction occurred to a very minor extent, there is still potential to impact assay results obtained given the extremely low detection levels. As such, the HLB plate was investigated to see if it generated more-reproducible results. As part of the development of the HLB method, wash solutions containing 5%, 10%, and 15% methanol were tested. The use of 10% and 15% methanol resulted in loss of the metabolite during the wash steps, so only one organic wash step using 5% methanol was used.
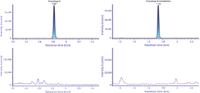
Figure 3: No detectable carryover was observed for both clopidogrel (upper limit of quantification [ULOQ] top left, blank bottom left) and the carboxylic acid metabolite (ULOQ top right, blank bottom right).
Because of the lower sensitivity of the HLB method compared to the mixed-mode cation exchange method, it was not possible to obtain the 1 pg/mL LLOQ using the previously established UHPLC–MS-MS conditions. The use of 1.0-mm i.d. (microbore) chromatography is known to give increases in sensitivity of up to fourfold over that obtained with a standard 2.1-mm column, thus a smaller diameter column was used. The UHPLC method also needed to be updated to accommodate the lower flow rate and the longer column re-equilibration time required for the 1-mm column. The resulting method showed reproducible results for the entire calibration range of 1–500 pg/mL.

Figure 4: Signal-to-noise values for clopidogrel (left) and carboxylic acid metabolite (right).
Assay Results
The resulting chromatograms obtained for standards clopidogrel and clopidogrel carboxylic acid, as well as the internal standards d4-clopidgrel and d4-clopidogrel carboxylic acid are shown in Figure 2. The metabolite and deuterated internal standard are eluted at 1.7 min, while clopidogrel and its internal standard are eluted at 2.8 min. The calibration curve was linear over the range of 1.00–500 pg/mL, with no carryover detected for either component (see Figure 3). The use of microelution SPE coupled with a 1.0-mm i.d. column gave rise to an LLOQ of 1 pg/mL for both clopidogrel and its metabolite. Clopidogrel showed a signal-to-noise ratio of 7:1 while the metabolite showed a signal-to-noise ratio of 5:1 (Figure 4). QC samples were injected in replicates of five at four different levels spanning the range of the calibration curve. All QCs met acceptance criteria of accuracy and precision ±20% for the LLOQ and accuracy and precision ±15% for all other QC levels (Table I).

Table I: QC statistical data for QC levels at LLOQ, low, mid, and high values
As previously stated in the method development discussion, another essential part of method reliability and robustness is the ability to separate the analytes of interest from any background interferences, such as choline-containing lipids. Comparison of the two MRM channels for clopidogrel and the acid metabolite with the precursor scan of 184 m/z over the mass range of 490–760 m/z is shown in Figure 5. As can be seen from the chromatogram, clopidogrel is eluted between two major regions of interference while the metabolite is eluted over 1 min before elution of the choline-containing lipids. Thus, successful separation of the components of interest from any potential impact of coeluted phospholipids was achieved.

Figure 5: Chromatographic resolution of clopidogrel and carboxylic acid metabolite from choline-containing phospholipids.
Conclusions
A systematic approach to the method development process was successfully carried out for clopidogrel and its inactive carboxylic acid metabolite. The optimal extraction method was determined to be by a solid-phase microelution extraction. The chromatographic conditions were then examined, leading to successful separation of clopidogrel and the carboxylic acid metabolite from coeluted phospholipids. This successful separation allowed for the development of a high sensitivity method for both components with an LLOQ of 1 pg/mL. A mock-validation accuracy and precision batch was run, and all standards and quality control samples met the required acceptance criteria put forth by the FDA. For each QC level, multiple replicates injected showed low variability between extracted samples. In addition, there was no detectable carryover for either compound.
References
(1) J.-M. Pereillo, M. Maftouh, A. Andrieu, M.-F. Uzabiaga, O. Fedeli, P. Savi, M. Pascal, J.-M. Herbert, J.-P. Maffrand, and C. Picard, Drug Metab. Dispos. 30, 1288–1295 (2002).
(2) A. Robinson, J. Hillis, C. Neal, and A.C. Leary, J. Chromatogr. B 848, 344–354 (2007).
(3) J.-J. Zou, H.-W. Fan, D.-Q. Guo, Y.-B. Li, S. Lin, Y.-B. Zhu, C.-X. Yu, J. Zhou, J.-H. Liu, and Y.-F. Hu, Chromatographia 70, 1581–1586 (2009).
(4) M. El Sadek, S. Moustafa, H. Kadi, A. Moneim, and A. Al-Hakami, Am. J. Anal. Chem. 2, 447–455 (2011).
(5) P.D. Rainville, N.W. Smith, D. Cowan, and R.S. Plumb, J. Pharm Biomed. Anal. 59, 138–150 (2012).
Jennifer L. Simeone is a senior scientist, Paul D. Rainville is a principal scientist, and Robert S. Plumb is a senior business manager with Waters Corporation in Milford, Massachusetts. Direct correspondence to: Jennifer_Simeone@waters.com.

Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.