Increasing Productivity of ADME Studies Using Accurate Mass Technology
Special Issues
A new time-of-flight mass spectrometer was evaluated for performing simultaneous metabolic stability measurement and metabolite identification with ultrahigh-pressure liquid chromatography. Six representative compounds (clomipramine, diclofenac, imipramine, haloperidol, verapamil, and midazolam) were incubated in rat liver microsomes at a more physiologically relevant substrate concentration (1 ?M). High-resolution full-scan and product-ion spectra were acquired in a single injection using generic methodology. Quantitative clearance of the parent was measured using the full-scan data. Major metabolites were identified using the accurate mass product ion spectra. High scanning speed allowed for a sufficient number of data points to be collected across the chromatographic peak for quantitative analysis. Sensitivity was sufficient for obtaining meaningful kinetics with a 1 ?M initial substrate concentration.
Increasing productivity in drug discovery and development continues to be a primary goal in pharmaceutical research. One of the most promising approaches to improving productivity is combining quantitative and qualitative analysis in a single analytical run. Quantitative information can be obtained on the parent compound while simultaneously acquiring qualitative information on metabolites in a completely automated fashion.
Metabolic stability studies in microsomes, a classic example of in vitro studies, have been used traditionally for estimation of hepatic clearance. For compounds exhibiting rapid metabolism, additional qualitative studies are performed to identify the major metabolites and identify metabolic "soft spots." This information aids the medicinal chemist in optimizing the structure. However, performing a separate analysis for metabolite identification is time-consuming.
A major improvement in productivity can be achieved if both quantitative clearance and qualitative soft spot analysis can be performed in a single analytical run. A key requirement is to obtain accurate mass product ion tandem mass spectrometry (MS-MS) spectra at high speed on a fast chromatographic time scale. In addition, due to the high-throughput nature of in vitro assays, hundreds of compounds must be analyzed weekly. Therefore, a generic data acquisition method without the need to optimize for individual compounds is highly desirable.
Due to their excellent sensitivity and high throughput, triple-quadrupole instruments have been the workhorse instrument for the quantitative portion of this application. Accurate mass instruments, such as time-of-flight (TOF) and orbital trapping analyzers, have been used for metabolite identification due to the high mass accuracy and resolution. Unfortunately, orbital trapping involves a compromise between resolution and speed. Traditional TOF technology has shown limitations in linearity and requires internal calibration to maintain mass accuracy. Modifying study designs, sample preparation procedures, or chromatography to accommodate instrument limitations is not acceptable.
The following details the experimental conditions and results using advanced accurate mass technology to combine quantitative and qualitative analysis for absorption, distribution, metabolism, and excretion (ADME) studies. Assay modifications are not necessary to realize the benefits of true integrated analysis in a single experiment.
Experimental Conditions
Sample Preparation: Six compounds (clomipramine, diclofenac, imipramine, haloperidol, verapamil, and midazolam) were incubated in rat liver microsomes at an initial substrate concentration of 1 µM in 96-well format. Standard high-throughput incubation conditions were used with time points of 0, 5, 15, 30, 60, and 120 min. The protein concentration was 1 mg/mL, and NADPH was present at 4 mM. The reaction was quenched using an equal volume of acetonitrile and then diluted 1:1 with water before analysis. The final substrate concentration at t = 0 was 0.25 µM.
Chromatography: Sample analysis was performed on a TripleTOF 5600 MS system (AB SCIEX, Foster City, California) coupled with a Prominence UFLC-XR high performance liquid chromatography (HPLC) system (Shimadzu, Columbia, Maryland). A generic acetonitrile–water–0.1% formic acid gradient was used on a 50 mm × 2 mm, 2.5-µm Phenomenex Synergi Polar-RP column (Phenomenex, Torrance, California). The total run time was 3.7 min, and the injection volume was 10 µL.
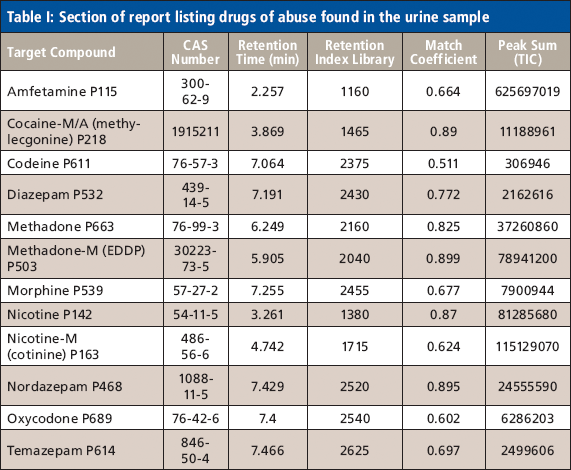
Mass Spectrometry: A completely generic method was used for data acquisition on all compounds and all samples. The method consisted of a TOF-MS survey scan followed by two IDA TOF MS-MS scans. The mass range was m/z 100–1000 for both MS and MS-MS. An accumulation time of 100 ms was used for each scan. Dynamic background subtraction was applied for IDA criteria and a collision energy of 35 eV with a spread of ±10 eV was used for the MS-MS scans. External mass calibration was performed automatically using a calibrant delivery system. Data processing was performed using proprietary software (MultiQuant and MetabolitePilot, AB Sciex, Figure 1).
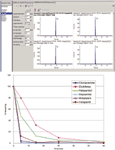
Figure 1: Data processing (MultiQuant software): (a) TOF-MS data were processed for multiple analytes to obtain quantitative data. (b) Metabolic stability profiles of six common substrates incubated at 1 µM and analyzed using TOF-MS with IDA triggered MS-MS.
Results
Sensitivity and Speed: The software was used to process the TOF-MS data and generate all quantitative information. An XIC window of ±10 mDa was used for all compounds. Peak areas were used to plot the % remaining relative to t = 0 (Figure 1b) and the software was used to process the data for metabolites.
Metabolic stability studies are best performed at a lower substrate concentration (1 µM), which is more physiologically relevant. Enough sensitivity is required to detect at least 1% of parent remaining to obtain meaningful kinetic data. The system demonstrated excellent sensitivity and speed. For example, 0.1 % of midazolam remaining was detected easily (Figure 2). This represents a concentration of 0.25 nM or 0.8 pg on column.
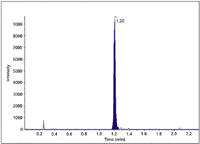
Figure 2: TOF-MS analysis of midazolam (t = 30 min) with 0.1% of parent remaining (initial concentration 0.25 µM at t = 0). XIC generated using a ±10 mDa window. Eight points were sampled across the peak in IDA.
Sensitivity does not mean compromising speed. A minimum of eight points was obtained in IDA mode (with two dependent MS-MS scans) across a 4-s peak while maintaining 30,000 resolution in both MS and MS-MS mode. In fact, each data point consisted of three scans (TOF-MS plus two MS-MS scans) for a total of 24 scans.
High Resolution and Mass Accuracy: Excellent mass accuracy and resolution were achieved in both MS and MS-MS scans (Figure 3). High mass accuracy in MS-MS greatly facilitates structure elucidation as it allows unambiguous assignment of elemental composition.
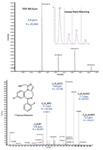
Figure 3: Mass accuracy and resolution in TOF-MS and MS-MS mode. Haloperidol parent in the 5-min time point is shown on the top with a near perfect match of isotope pattern (inset). Product ion spectrum of hydroxy midazolam in the 5-min time point acquired with IDA is shown on the bottom.
Using the software described here, data from multiple samples and compounds was processed unattended in batch mode. The software reported metabolites detected and elemental compositions in a concise and powerful yet user friendly interface.
Excellent separation, metabolite coverage, and speed were demonstrated using a generic IDA method (Figure 4).
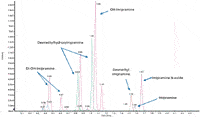
Figure 4: Broad coverage for Phase I metabolites. Overlay of accurate mass XICs for imipramine incubation at 5 min. A wide range of imipramine Phase I metabolites were detected using a generic TOF-MS IDA method in under 2.5 min.
No Compromises: Successful application of quant/qual technology to in vitro ADME was demonstrated using the system described here. No compromises were made in resolution to achieve speed or sensitivity. A relatively low substrate concentration of 1 µm was used successfully. Standard incubation and HPLC conditions were used with no need to make any modifications in order to gain the benefits of quant /qual.
Conclusion
The combination of speed, sensitivity, mass accuracy, and resolution enables routine quant/qual analysis. For metabolic stability, we have shown the ability to perform parent quantification, metabolite detection, and obtain accurate mass MS-MS information all in a single run.
Acknowledgments
The authors would like to thank Drs. Christie Hunter and Tanya Gamble for their invaluable contributions and review.
Hesham Ghobarah, Suma Ramagiri, and Jim Ferguson are application scientists with AB Sciex, Toronto, Canada, and Framingham, Massachusetts.
References
(1) R.C. King, R.Gundersdorf, and C.L. Fernández-Metzler, Rapid Commun Mass Spectrom. 17, 2413–2422 (2003).
(2) A.C. Li, D. Alton, M.S. Bryant, and W.Z. Shou, Rapid Commun Mass Spectrom. 19, 1943–1950 (2005).
(3) C.E.C.A. Hop, M.J. Cole, R.E. Davidson, D.B. Duignan, J. Federico, J.S. Janiszewski, K. Jenkins, S. Krueger, R. Lebowitz, T.E. Liston, W. Mitchell, M. Snyder, S.J. Steyn, J.R. Soglia, C. Taylor, M.D. Troutman, J. Umland, M.West, K.M.Whalen, V. Zelesky, and S.X. Zhao, Curr Drug Metab. 9, 847–853 (2008).
(4) J.D. Gilbert, T.V. Olah, and D.A. McLoughlin, Biochem. Biotechnol. Appl. Electrospray Ion. Mass Spectrom. 619, 330–350 (1996).
(5) J.B. Houston, Biochem Pharmacol. 47, 1469–1479 (1994).

Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.