Accurate Mass: Why It's the Best Solution for Metabolite Identification in Discovery, Development, and Clinical Applications
Special Issues
Accurate-mass approaches offer a significant advance over nominal-mass approaches in the arena of qualitative analysis, and some of the analytical approaches can now be conducted in a relatively routine manner.
Accurate mass offers a significant advance over nominal-mass systems for metabolite studies. Accurate-mass data provide an increased ability to identify the metabolites observed through enhanced parent and fragment ion specificity. Examples provided show how accurate-mass data can differentiate between analytes of interest and the matrix. These examples also demonstrate how accurate-mass data of nonradiolabeled samples can address the challenges of in vivo metabolite identification studies previously only attempted with the combination of liquid chromatography–mass spectrometry and flow scintillation analysis. Further, a discussion of the use of accurate mass in glutathione profiling exemplifies the advantages of using accurate mass for in vitro applications.
In recent years, accurate mass–based analyses have become an increasingly important approach for qualitative mass spectrometry (MS) analyses (1–6). The use of accurate-mass applications has steadily replaced the use of nominal-mass approaches (7–11), and many scientists who are new to accurate-mass instrumentation are making greater use of the technologies now available.
The goal of this paper is twofold. First, it describes the theoretical basis of why accurate-mass approaches offer a significant advance over nominal-mass approaches in the arena of qualitative analysis, and specifically metabolite identification. Second, it describes some of the analytical approaches that can now be conducted in a relatively routine manner — analytical approaches that were far from routine using the nominal-mass instrumentation.
Definitions
We should begin the discussion of accurate mass–based analyses with some key definitions, provided below.
- Average mass is the mass calculated using the average atomic mass of each element weighted for its natural isotope abundance.
- Nominal mass is the integer mass of the most naturally occurring stable isotope.
- Monoisotopic mass is the calculated mass of an ion or molecule calculated using the mass of the most abundant naturally occurring isotope of each element.
- Exact mass is the calculated mass of an ion or molecule containing a single specified isotope of each atom.
- Accurate mass is the measured mass of an ion measured to high mass accuracy.
- High-resolution mass spectrometry (HRMS) is an MS technology that exhibits high mass resolving power.
The average mass is a value that is frequently used by medicinal chemists, but has no meaning in MS. No molecule exists that is represented by the average mass; the various isotopes present in a molecule generate an isotope pattern in the mass spectrum. The terms monoisotopic mass and exact mass are often used interchangeably because the most abundant naturally occurring isotope commonly is used in calculating an exact mass.
Resolution
Unit mass resolution describes the capability of a mass spectrometer to differentiate between adjacent masses such as m/z 50 and 51 or m/z 1000 and 1001. This term is appropriate when discussing the resolution of a quadrupole or ion-trap mass spectrometer.
The definition of resolving power that is appropriate for quadrupole orthogonal acceleration time-of-flight (QTOF) and orbital trap mass spectrometers is based on full width at half maximum (FWHM). Thus, resolving power is defined as the mass being analyzed divided by the peak width of the raw mass data at that mass. So a mass spectrometer that detects an ion at m/z 500 that has a peak width of 0.1 Da is operating at a resolving power of 500/0.1, or 5000. A mass spectrometer exhibiting a resolving power of 5000 can differentiate between m/z 50.000 and 50.010 or m/z 1000.000 and 1000.200.
Because mass resolution is a function of the mass of the ion being examined, when an instrument resolution is stated, the mass at which that resolution was determined also must be defined. In the example above, the mass spectrometer is exhibiting a resolution of 5000 at mass 500.
Mass Accuracy
Mass accuracy is a key parameter of mass spectrometer performance and determines the specificity of the mass measurement being made. The most commonly used definition of mass accuracy is given in parts per million (ppm) and is determined as the difference between an exact (or theoretical) and measured (accurate) mass. Table I displays the ppm errors for three different measured masses that have the same absolute mass error (25 mDa) relative to their exact masses. The ppm error, because it is a relative term, varies with the mass of the ion being examined. Traditionally, a mass error of 5 ppm has been accepted by medicinal chemistry journals as sufficient to provide a definitive elemental composition. Because ppm mass accuracy is a relative term, however, mDa mass accuracy, which is as an absolute measurement of instrument performance, often is used as a convenient day-to-day tool.
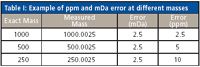
Table I: Example of ppm and mDa error at different masses
A mass spectrometer with high mass resolving power generally is considered an essential tool for achieving good mass accuracy. In reality, high resolving power is used to eliminate chemical interferences and is not otherwise required for obtaining good mass accuracy. In fact, accurate-mass measurements can be achieved routinely with even a quadrupole mass spectrometer operated at low mass resolving power if interferences are not a problem. However, given that the samples being analyzed usually are not pure standards but rather are mixtures or chromatographic peaks from LC–MS analyses, interferences are a frequent occurrence and HRMS is required to obtain quality accurate-mass data.
Why Accurate Mass?
The application of accurate-mass technology offers several advantages in conducting metabolite identification studies when compared with the use of nominal-mass instrumentation such as triple-quadrupole or ion-trap mass spectrometers.
A review of the three main goals in conducting metabolite identification studies creates a framework for understanding these advantages. These goals are the same whether the samples are derived from in vitro or in vivo experiments.
The first goal of metabolite identification is to differentiate between ions derived from the drug itself and drug-related metabolites and ions derived from the matrix. The second goal is to identify the biotransformation that has taken place, such as addition of an oxygen atom or loss of a methyl group. The third goal is to define which site of the drug molecule has undergone biotransformation. Accurate-mass data enable all three of these goals to be achieved more quickly than when using nominal-mass data.
A paradigm shift is possible in using accurate-mass approaches because the data are rarely compromised with interferences and thus provide more information about the molecule being analyzed. Compared to nominal-mass studies, the overall result is that conclusions are reached more rapidly, with a higher level of confidence in their accuracy.
There are four main reasons that accurate-mass data are superior to nominal-mass data in conducting metabolite identification studies, and these are discussed below.
Analyte Versus Matrix Specificity
How is it that having accurate-mass data enables differentiation between the analytes of interest and the matrix? In most instances, the matrix components to be eliminated as false positives are derived from biological sources such as microsomes, hepatocytes, plasma, or urine. Such components are predominantly made of carbon, hydrogen, nitrogen, and oxygen.
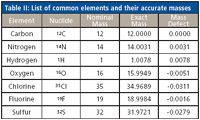
Table II: List of common elements and their accurate masses
Most xenobiotics that are being analyzed, though mainly derived of the same elements, often contain a higher level of unsaturation and occasionally incorporate halogens. Table II lists some basic parameters of common elements. Carbon does not contribute to the fractional mass (the numerals after the decimal point) of a molecule's mass, whereas both hydrogen and nitrogen increase the fractional mass, and oxygen, halogens, and sulfur decrease the fractional mass. Thus, the fractional mass of a molecule will change with the elements that constitute the structure of the compound.
For example, for each double bond introduced into a molecule, the fractional mass will decrease by 0.0156 Da. Because many of the xenobiotics that are drug candidates are more highly aromatic than naturally occurring "interferences" such as peptides, the observed fractional mass is lower. A stylized example of this is shown in Figure 1, in which a tripeptide, Tyr-Gly-Gly, is compared with a possible drug compound of similar nominal mass. The mass difference of 32 mDa would be easily differentiated using a modern accurate-mass instrument.
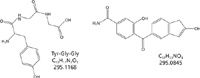
Figure 1: Example of a tripeptide and a possible xenobiotic with similar nominal mass.
The difference in fractional mass enables elimination of false positive mass "hits" when dealing with biological matrices. Figure 2 reproduces two extracted ion chromatograms from the same data file. The top trace uses a mass window of ±0.5 Da and the bottom trace uses a mass window of ±0.005 Da. The nominal-mass top trace has multiple peaks or "hits" for the parent compound, whereas the accurate-mass bottom trace has only one peak.
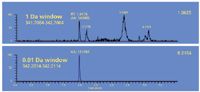
Figure 2: Two extracted ion chromatograms from the same data file. The top trace is generated using a 1-Da wide window. The bottom trace is from using a 10-mDa wide window.
Parent Ion Specificity
Accurate-mass data facilitate differentiation of biotransformations that result in an identical nominal-mass shift. For example, when using a nominal-mass instrument, it is impossible to differentiate between M + O – 2H or M + CH2 because the mass shift (in both instances) is +14 Da. When the same comparison is made using accurate-mass data, the difference is very clear, because M + O – 2H (M + 13.9793) or M + CH2 (M + 14.0156) results in a mass shift that differs by 36 mDa.
Fragment Ion Specificity
An essential step in conducting a metabolite identification study is to determine which fragment ion in the MS-MS spectrum is derived from the correct substructural moiety of the parent molecule. This information is used to compare the MS-MS spectra of a metabolite with the parent drug and thus determine the part of the molecule that has undergone biotransformation. The use of accurate-mass data in this step can prevent errors in localization of the biotransformation, because nominal-mass data cannot differentiate between different substructures in the molecule that provide the same nominal-mass fragment ions.
Figure 3 shows a molecule that would provide fragment ions with similar nominal mass that would require an accurate-mass analysis to be correctly interpreted. Incorrectly interpreting the MS-MS spectrum of the parent drug would result in incorrect localization of the biotransformation. An incorrect localization of the site of metabolism could lead to synthetic chemisty decisions to address a metabolic instability that would alter the wrong site on the molecule.
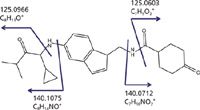
Figure 3: Example of a potential drug molecule that provides multiple fragment ions with the same nominal but different exact masses.
Analytical Sensitivity
Nominal-mass instruments use MS-MS methods to obtain specificity, such as multiple reaction monitoring (MRM) for quantitative assays and precursor ion scans and constant neutral loss scans for qualitative screening or identification. These MS-MS approaches are less sensitive than full-scan experiments, simply as a consequence of transmission efficiency in such MS-MS instrument modes. The application of accurate mass using full-scan experiments is more sensitive than nominal-mass MS-MS methods because equal, or superior, specificity is obtained without the resultant loss of ion current.
The Role of Metabolite Identification in Various Stages of Drug Development
Metabolite identification plays an important role throughout the discovery, preclinical, and clinical stages of small-molecule drug development (see Figure 4).

Figure 4: Schematic of different metabolite identification experiments performed throughout the drug discovery, preclinical, and clinical stages of drug development.
In drug discovery, metabolite identification can be used to determine the routes of excretion of a drug and its metabolites, to define the sites on a molecule that are subject to metabolism, to assess the potential for formation of reactive metabolites, and to compare the metabolites formed across various species, including humans.
In preclinical drug development, in vivo studies, typically in rats, dogs, and monkeys, are designed to confirm that the metabolite profile observed across species in the in vitro studies are also observed in vivo. The mass balance study also illuminates the routes of excretion and their relative quantitative importance.
The clinical development role of metabolite identification is generally twofold. First, it determines that the circulating metabolites observed in humans were also present in the toxicity studies filed in the investigational new drug (IND) application, thus demonstrating that the toxicity of the human metabolites has been tested. Second, it defines the structure of the metabolites observed in human plasma, urine, and feces and the relative importance of the observed route of excretion.
Three applications areas of metabolite identification are discussed below, highlighting the advantages that using accurate mass provides.
Routes of Excretion
Metabolic stability assays play an important role in modifying compound structures to improve the pharmacokinetic characteristics of potential drug molecules. Microsomes are useful if phase I metabolism is the major route, and hepatocytes are needed if phase II is an important pathway. Just using hepatocytes is a possible, albeit an expensive, default position to take. Conducting a bile duct cannulated study (typically in rat) to define the routes of elimination is an option when applying modern accurate-mass instrumentation. These studies can be carried out using nonlabeled drug because of the analytical specificity offered by the use of HRMS, so meaningful data can be obtained from bile, urine, or plasma.
Figure 5 shows data from the bile of a rat dosed intravenously with verapamil. The three extracted ion chromatograms are for verapamil itself, demethylated verapamil, and the glucuronide conjugate of the demethylated metabolite. The glucuronide metabolite represents approximately 20% of the drug-related material and the demethylated metabolite accounts for approximately 25% of the material. Thus, hepatocytes would be the best model for conducting metabolic stability studies with verapamil and its analogs.
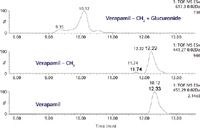
Figure 5: Extracted ion chromatograms of bile from a rat dosed with verapamil. The top trace is the glucuronide of the demethylated metabolite, the middle trace is the demethylated metabolites, and the bottom trace is verapamil itself.
Metabolites in Safety Testing
Both the US Food and Drug Administration and the European Medicines Agency require pharmaceutical companies to identify important metabolites that are observed circulating in human plasma, to ensure that the circulating human metabolites have had their toxicological effects tested in appropriate animal studies (12,13). This requirement means that studies must be conducted to demonstrate that the same metabolites that are observed in humans are also observed in at least one of the safety species at similar or greater abundance than in humans. Use of accurate-mass approaches enables studies to be conducted that meet these regulatory requirements using the preclinical and clinical studies that are a normal part of drug development.
Figure 6 reproduces data from human, rat, and dog plasma samples generated by proportional pooling of samples collected over a 24-h period (14). This method provides a single sample for each species that was representative of exposure of drug-related material over that time period. The extracted ion chromatograms depicted provide data on the unchanged drug and its mono-oxygenated metabolite observed at 7.4 and 6.6 min, respectively. The relative abundance of the M + 16 metabolite and the parent drug are similar in rat and human; thus, if the rat provided safety coverage for the parent drug, at this dose, it would also provide safety coverage for the M + 16 metabolite.

Figure 6: Extracted ion chromatograms from human, dog, and rat plasma showing the drug (tR 7.4 min) and the mono-oxygenated metabolite (tR 6.6 min). (Adapted from reference 2.)
A potential concern in metabolite safety studies is the possibility of a metabolite accumulating to a different degree upon multiple dosing of the parent drug. Comparing the proportional pooled sample from the first day of dosing to a proportional pooled sample from a later, steady-state time period enables an assessment of the degree of accumulation of the drug and its various metabolites. This comparison is demonstrated in Figure 7, where the parent drug and a metabolite are observed at 14.7 and 8.5 min, respectively. The pharmacokinetic (PK) data for these samples indicate that the area under the concentration time curve from 0 h to 12 h (AUC0–12 h) increases from 30.2 on day 1 to 57.9 on day 10. In the two chromatograms, at day 1 the parent peak is 186 counts and doubles to 384 on day 10, and the metabolite peak also doubles from 12 on day 1 to 23 on day 10. Thus, the proportional pooled sample data are in complete agreement with the clinical pharmacokinetic data. Establishing that a metabolite (disproportionately) accumulates provides early information that would indicate a potential need for drug–drug interaction studies, allowing these studies to be incorporated into the clinical development program. Having this information early in the clinical development of a drug provides time to synthesize the metabolite and to have an appropriate standard available for quantitative monitoring if necessary.

Figure 7: Comparison of day 1 and day 10 human plasma, showing the accumulation of parent drug (tR 14.7 min) and metabolite (tR 8.5 min). (Adapted from reference 2.)
In both in vivo examples (routes of excretion and metabolites in safety testing), the use of accurate-mass approaches in combination with the use of nonlabeled drug offers a less costly and equally effective way of obtaining metabolite identification data from complex biological samples. When using nominal-mass instrumentation in the past, effective analysis of in vivo animal samples required the synthesis of radiolabeled drug, and dosing a labeled drug to humans required the production of a standard qualified according to good manufacturing practices, both costly endeavors. The most obvious advantages of using accurate-mass approaches are seen in the current ability to analyze in vivo samples dosing cold drug and still obtain the same level of confidence in the data as when using labeled drug.
Reactive Metabolites
The metabolic bioactivation of drugs to reactive intermediates is a potential cause of clinical toxicities. Covalent binding assays provide a direct quantitative measure of the amount of reactive intermediates formed; however, such an assay requires the use of costly and time-consuming radiolabeled compound.
The use of trapping agents is an alternative method to study reactive intermediates. Glutathione (GSH), a natural detoxification reagent, is one of the most effective nucleophilic trapping agents because its conjugates are stable chemical species that are amenable to detection by LC–MS. GSH adduct screening in drug discovery can detect the potential for toxicity of drug candidate as a result of covalent binding.
There are three main approaches using nominal-mass instrumentation that are applied to GSH adduct screening: neutral loss scanning (monitoring loss of 129 Da) (15), parent ion scanning (monitoring 272 Da in negative ion mode) (16), and predicted MRM scanning (17).
Neutral loss scanning offers good selectivity and is an easy experiment to perform; its downsides include low analytical sensitivity, data containing false positives and false negatives, and up to 30% of adducts being missed. The false negatives occur because no doubly charged GSH adducts will provide a signal when monitoring the loss of 129 Da, and when other cleavages occurring in the molecule are predominant, the neutral loss of 129 may not be generated.
Parent ion scanning of the ion 272 Da in negative ion mode offers reasonable selectivity, and like neutral loss scanning is an easy experiment to conduct, but the data contain both false positives and false negatives, because other cleavages in the molecule predominate and the 272 Da ion may not be formed.
Predicted MRM scanning is both selective and sensitive, but each molecule requires a new set or MRM transitions to be defined, making it a tedious experiment to set up. This approach is also subject to false negative findings because no doubly charged GSH adducts will provide a signal when monitoring the MRM transitions based on the parent mass. In addition, when other cleavages in the molecule are predominant, the cleavages used to define the MRM transitions based on the parent cleavage will not be observed.
The use of full-scan accurate-mass approaches (15–17) has several advantages. Experiment setup is easy, and the methods are more sensitive because no collision-induced dissociation step required. The methods provide high specificity as a result of the use of accurate-mass data. Lastly, these approaches provide full coverage for doubly charged GSH adducts.
These advantages make the application of accurate-mass approaches the best choice in all such screening methods for small molecule qualitative analysis. This is shown in Figure 8, which depicts a stylized molecule with a GSH adduct, with the white box demonstrating the unpredictable cleavages that limit the utility of the neutral loss, parent ion, and MRM approaches. The red box and yellow brackets denote the part of the GSH adduct that is the focus of the parent ion and neutral loss methods, respectively.

Figure 8: Stylized drug with a glutathione (GSH) adduct, showing potential unanticipated fragments (white box) and the part of the GSH that is monitored in parent ion (red box) and neutral loss (yellow brackets) scanning.
Conclusions
The three examples discussed in this article exemplify the reasons why modern accurate-mass approaches are superior to nominal-mass approaches: speed, sensitivity, reduced cost, reduction or elimination of false positives or false negatives, and higher confidence in resultant conclusions. This enhanced specificity and sensitivity, routinely observed in conjunction with easy experimental setup, makes accurate mass the first choice for conducting metabolite identification experiments at any stage of drug discovery or development.
Philip R. Tiller, PhD, is the vice-president of RMI Laboratories in North Wales, Pennsylvania. He can be reached at (215)699-8871 or philip_tiller@rmilaboratories.com.
References
(1) K. Bateman, J. Castro-Perez, M. Wrona, J.P. Shockor, K. Yu, R. Oballa and D.A. Nicoll-Griffith, Rapid Commun. Mass Spectrom. 21, 1485–1496 (2007).
(2) P.R. Tiller, S. Yu, K. Bateman, J. Castro-Perez, I.S. Mcintosh, Y. Kuo, and T.A. Baillie, Rapid Commun. Mass Spectrom. 22, 3510–3516 (2008).
(3) P.R. Tiller, S. Yu, J. Castro-Perez, K.L. Filgrove, and T.A. Baillie, Rapid Commun. Mass Spectrom. 22, 1053–1061 (2008).
(4) H. Zhang, D. Zhang, K. Ray, and M. Zhu, J. Mass Spectrom. 44, 999–1016 (2009).
(5) M. Zhu, M. Li, D. Zhang, K. Ray, W. Zhao, W.G. Humphreys, G. Skiles, M. Sanders, and H. Zhang, Drug Metab. Dispos. 34, 1722 (2006)
(6) H. Zhang, M. Zhu, K.L. Ray, L. Ma, and D. Zhang, Rapid Commun. Mass Spectrom. 22, 2082 (2008).
(7) S.K. Chowdhury (Ed.), "Identification and Quantification of Drugs, Metabolites and Metabolizing Enzymes by LC-MS," in Progress in Pharmaceutical and Biomedical Analysis, vol. 6 (Elsevier, Amsterdam, 2005).
(8) R.F. Staack and G. Hopfgartner, Anal. Bioanal. Chem. 388, 1365 (2007).
(9) C. Prakash, C.L. Shaffer, and A. Nedderman, Mass Spectrom. Rev. 26, 340 (2007).
(10) S. Ma and R. Subramanian, J. Mass Spectrom. 41, 1121 (2006).
(11) M. Yao, L. Ma, W.G. Humphreys, and M. Zhu, J. Mass Spectrom. 43, 1364 (2008).
(12) U.S. Food and Drug Administration, Guidance for Industry. Safety Testing of Drug Metabolites (US FDA, Rockville, Maryland, February 2008).
(13) International Conference on Harmonization. M3(R2), Guidance on Nonclinical Safety Studies for the Conduct of Human Clinical Trials and Marketing Authorization for Pharmaceuticals. (Geneva, Switzerland, June 2009).
(14) L.E. Riad, K.K. Chan, and R.J. Sawchuk, Pharm. Res. 8, 541–543 (1991).
(15) T.A. Baillie and M.R. Davis, Biol. Mass. Spectrom. 22, 319–325 (1993).
(16) C.M. Dieckhaus, C.L. Fernandez-Metzler, R. King, P.H. Krolikowski, and T.A. Baillie, Chem. Res. Toxicol. 18, 630–638 (2005).
(17) J. Heng, L. Ma, B. Xin, T. Olah, W.G. Humphreys, and M. Zhu. Chem. Res. Toxicol. 20, 757–766 (2007).

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
The Fundamental Role of Advanced Hyphenated Techniques in Lithium-Ion Battery Research
December 4th 2024Spectroscopy spoke with Uwe Karst, a full professor at the University of Münster in the Institute of Inorganic and Analytical Chemistry, to discuss his research on hyphenated analytical techniques in battery research.
Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.