Surface-Enhanced Nanosensors
January 2007. This review article summarizes biological applications that utilize surface plasmon resonance, localized surface plasmon resonance, and surface-enhanced Raman spectroscopy.

Widespread interest in surface-enhanced spectroscopy has been rekindled by the development of new fabrication methods for plasmonic materials (1,2), theoretical modeling (3,4), spectroscopic instrumentation (5–9), and novel detection schemes (10). When the incident photon frequency is resonant with the collective oscillation of the conduction electrons, the local electromagnetic fields near the surface of the plasmonic nanostructure are enhanced. The enhancement in the electromagnetic fields is responsible for the intense signals observed in all surface-enhanced spectroscopies, such as surface plasmon resonance (SPR) spectroscopy (6,7,11,12), localized surface plasmon resonance (LSPR) spectroscopy (13,14), and surface-enhanced Raman spectroscopy (SERS) (15,16). This paper summarizes biological applications that utilize the aforementioned surface-enhanced spectroscopies: comparative study of surface plasmon and localized surface plasmon resonance sensor using concanavalin A as a model protein, development of an ultrasensitive nanoscale optical biosensor for the detection of cytochrome P450, and fabrication of a highly sensitive and stable surface-enhanced Raman spectroscopy sensor for detection of an anthrax biomarker.
Experimental
Materials: Ag (99.99%) was purchased from D.F. Goldsmith (Evanston, Illinois). Glass substrates were 18 mm diameter, No. 2 cover slips from Fisher Scientific (Pittsburgh, Pennsylvania). Surfactant-free white carboxyl-functionalized polystyrene latex nanospheres with diameters of 280, 390, 450, 510, and 590 nm were obtained from Interfacial Dynamics Corporation (Portland, Oregon) or Duke Scientific (Palo Alto, California). Tungsten vapor deposition boats were purchased from R.D. Mathis (Long Beach, California). Water (18.2 MW/cm) was obtained from an ultrafilter system (Milli-Q, Millipore, Marlborough, Massachusetts). All of the other chemicals, reagents, and solvents were purchased from Aldrich Chemical (Milwaukee, Wisconsin) or Fisher Scienti1/2c (Fairlawn, New Jersey) without further purification.
Thin layer chromatography (TLC) analyses were performed on aluminum-backed aluminum oxide 60 F-254 neutral with a 0.2-mm layer thickness (type E, E. Merck, Darmstadt, Germany) in 10:1 (v/v) hexanes–ethyl acetate.
Calcium dipicolinate (CaDPA) was prepared from DPA and calcium hydroxide according to the method of Bailey and colleagues (69).
B. subtilis spore samples were prepared according to the previously published method (29). Approximately 1 g of sample was determined to contain 5.6 × 1010 spores by optical microscope measurements (data not shown). The spore suspension was made by dissolving spores in 0.02 M HNO3 solution and by sonicating for 10 min, which effectively extracts CaDPA from spores. This concentration of the HNO3 solution was selected because of its capability for CaDPA extraction and its benign effect on the AgFON SERS activity. The sonication procedure was performed because no SERS signal of CaDPA was observed from the spore solution prior to sonication (data not shown).
AgFON substrate fabrication: Glass substrates were pretreated in two steps: piranha etch (Caution: piranha solution should be handled with great care), 3:1 H2SO4–30% H2O2 at 80 °C for 1 h, was used to clean the substrate; and base treatment, 5:1:1 H2O–NH4OH–30% H2O2 with sonication for 1 h, was used to render the surface hydrophilic. Approximately 2 mL of the nanosphere suspension (4% solids) was drop-coated onto each substrate and allowed to dry in ambient conditions. The metal films were deposited in a modified Consolidated Vacuum Corporation vapor deposition system with a base pressure of 10-6 torr. The deposition rates for each film (10 Å/s) were measured using a Leybold Inficon XTM/2 quartz crystal microbalance (East Syracuse, New York). AgFON substrates were stored in the dark at room temperature before use.
Atomic layer deposition (ALD): Alumina films were fabricated on the Ag nanoparticles by ALD. The reactor utilized in these experiments is similar to that mentioned in previous publications (54). Trimethyl aluminum (TMA) and deionized H2O vapors were alternately pulsed through the reaction chamber utilizing N2 as the carrier gas at a mass flow rate of 360 sccm and a pressure of 1 torr using a growth temperature of 50 °C. One complete ALD cycle takes 42 s and includes four steps: TMA reactant exposure time = 1 s; N2 purge following TMA exposure time = 10 s; H2O reactant exposure time = 1 s; and N2 purge following H2O exposure time = 30 s. Long purge times are necessary at low temperatures to prevent chemical vapor deposition of alumina (72,73). A previous study indicated nearly ideal layer-by-layer growth of the ALD alumina on Ag surfaces with an average rate of ~2 Å/cycle (54). This result greatly simplifies the interpretation of the thickness of the alumina overlayers, which can be deduced easily from the number of ALD cycles.
Scanning electron microscopy (SEM): SEM images of alumina-coated AgFON were observed with a Hitatchi S-4700-II SEM (Hitachi, Schaumburg, Illinois).
LSPR reflectance spectroscopy: Reflectance measurements were carried out using an SD2000 spectrometer coupled to a reflection probe (Ocean Optics, Dunedin, Florida) and a halogen lamp (F-O-Lite H, World Precision Instruments, Sarasota, Florida). The reflection probe consists of a tight bundle of 13 optical fibers (12 illumination fibers around one collection fiber) with a usable wavelength range of 400–900 nm. All reflectance spectra were collected against a mirrorlike Ag film over glass surface as a reference in air.
SERS apparatus: The macro Raman system consists of an interference bandpass filter (Coherent, Santa Clara, California), a 1-in. long pass dielectric edge filter (Sermrock, Rochester, New York), a single-grating monochromator with the entrance slit set at 100 mm (model VM-505, Acton Research Corporation, Acton, Massachusetts), a liquid-nitrogen-cooled CCD detector (model Spec-10:400B, Roper Scientific, Trenton, New Jersey), and a data acquisition system (Photometrics, Tucson, Arizona). A compact diode laser (model RL785, Renishaw plc, United Kingdom) was used to generate 785 nm. All the measurements were performed in ambient conditions.
Surface Plasmon Resonance and Localized Surface Plasmon Resonance
SPR reflectivity measurements can be used to characterize the thickness or refractive index of ultrathin organic and biopolymer films at noble metal surfaces. Since the introduction of the Biacore SPR instrument (6,7), SPR spectroscopy has become widely used in the fields of chemistry and biochemistry to characterize biological surfaces and to monitor binding events. LSPR spectroscopy is a noble metal nanoparticle-based optical sensing technique, effective for quantitative detection of chemical and biological targets (13,14,17,18). Sensing is accomplished by monitoring the wavelength shift in the LSPR extinction or scattering maximum (λmax) induced by the binding of target analytes to the nanoparticle surface. The concentration of target analytes is quantitatively related to the shift in λmax. LSPR nanosensors have been demonstrated as sensitive platforms for the detection of streptavidin (17), antibiotin (18), concanavalin (14), Alzheimer's disease biomarkers (19), and many other biorecognition events (20).
Recently, a comparative analysis of the properties of two optical biosensor platforms, the propagating SPR sensor based upon a planar, thin-film gold surface, and the LSPR sensor based upon surface-confined Ag nanoparticles fabricated by nanosphere lithography (NSL), was presented (14). The binding of concanavalin A (ConA) to mannose-functionalized self-assembled monolayers was chosen to highlight the similarities and differences between the responses of the real-time angle shift SPR and wavelength shift LSPR biosensors. During the association phase in the real-time binding studies, both SPR and LSPR sensors exhibited qualitatively similar signal-versus-time curves (Figure 1). However, in the dissociation phase, the SPR sensor showed an approximately five times greater loss of signal than the LSPR sensor (Figure 1). A comprehensive set of nonspecific binding studies demonstrated that this signal difference was not the consequence of greater nonspecific binding to the LSPR sensor. To understand the LSPR realtime response of the ConA binding to the mannose-functionalized surface, Ag nanoparticles with 16-, 25-, and 50-nm out-of-plane heights were constructed. The Ag nanoparticles with larger aspect ratios showed larger dissociation responses than Ag nanoparticles with smaller aspect ratios. In addition, theoretical modeling indicated that the long range of the electromagnetic fields surrounding Ag nanoparticles with large aspect ratios showed a greater dissociation response than Ag nanoparticles with smaller aspect ratios. Although the SPR sensors are more sensitive to large molecules, a huge signal due to nonspecific binding of the molecules is also inherent to the SPR sensors. In contrast, to minimize the signal due to nonspecific binding of the molecules in the LSPR sensor, the height of the nanoparticles can be tuned easily with respect to the size of binding molecules of interest.

Figure 1
Resonance Surface Plasmon Spectroscopy: Low Molecular Weight Substrate Binding to Cytochrome P450
When analytes are optically transparent, the observed LSPR shift is only weakly dependent upon the LSPRmax of the nanoparticles (17–19,21). Because many biomolecules contain visible chromophores, it is important to broaden the scope of LSPR sensing by exploring electronically resonant adsorbates in biosensing events. When resonant molecules are adsorbed on nanoparticles, the induced LSPR shift is found to be strongly dependent upon the spectral overlap between the electronic resonance of the adsorbates and the plasmon resonance of the nanoparticles (22). Specifically, a large red shift occurs when the nanoparticles' LSPR is located at a slightly longer wavelength than the adsorbate's molecular resonance wavelength (that is, a factor of 3 greater than when the LSPR is distant from the molecular resonance) (22,23). This resonant LSPR response opens up the possibility of detecting the binding of a low molecular weight analyte to a protein receptor adsorbed on a nanoparticle. A proof-of-concept experiment for the binding of camphor (C10H16O, molecular weight = 152.24 g/mol) to cytochrome P450cam protein (CYP101) has been demonstrated (23). This system was selected because the electronic structure changes that occur when substrate binds have been well characterized (24,25). Indeed, this is the first demonstration that the binding of a small molecule (camphor) to a protein, CYP101(Fe3+ ), can generate an LSPR spectral change (23). Amplified spectral response to substrate binding is achieved when the LSPR of the silver nanosensor is optimized to be close to the molecular resonance of the protein. This study demonstrates that strong coupling between the molecular resonance and the intrinsic LSPR of the nanoparticles results in an amplified LSPR shift that is modulated by substrate binding, providing further insight into possible uses of plasmon resonance spectroscopy. Application of this finding to the screening for inhibitors of human cytochrome P450s is under investigation based upon these results.
Figures 2a and 2b show two representative sets of LSPR spectra for each step of the nanoparticle functionalization at off CYP101(Fe3+ ) resonance (Figure 2a) and close to CYP101(Fe3+ ) resonance (Figure 2b). To further study the wavelength-dependent relationship between the LSPR shifts and λmax,SAM, experiments were conducted to measure the LSPR response of nanoparticles while varying the initial LSPR wavelength (shown in Figure 2c). When the LSPR of the nanoparticles is located at a wavelength distant from the cytochrome P450 resonance, an average of ~19 nm red-shift is observed upon cytochrome P450 binding to the nanoparticles, and an ~6-nm blue-shift is observed upon camphor binding. However, this response is significantly amplified ~3–5 times when the LSPR of the nanoparticles is located at a slightly longer wavelength than the cytochrome P450 resonance (a 66.2-nm red-shift upon cytochrome P450 binding and a 34.7-nm blue shift upon camphor binding). It is observed for the first time that the binding of the substrate molecules to the protein receptor induces a blue shift in the LSPR of the nanosensors. The coupling between the molecular resonance of the substrate-free and substrate-bound cytochrome P450 proteins and the nanoparticles' LSPR leads to a highly wavelength-dependent LSPR response. This is the first example of the detection of small molecules binding to a protein-modified nanoparticle surface based upon LSPR. It is foreseeable that this discovery will provide guidance to the design and optimization of refractive index-based sensing for biological targets with resonant chromophores.

Figure 2
Surface-Enhanced Raman Spectroscopy
Compared with the refractive index-based detection schemes, SERS is a vibrational spectroscopic method that yields unique vibrational signatures for small molecule analytes, as well as quantitative information. The SERS signal transduction mechanism (26) has many characteristics that can be exploited in biosensing applications: sensitivity, selectivity, low laser power, and no interference from water molecules (27). Examples are trace analysis of DNA (28), bacteria (29–31), glucose (32–34), living cells (27,35), posttranslational modification of proteins (36), enzyme (37), chemical warfare agents (38), and carbon nanotubes (2,39,40). A miniaturized, inexpensive, and portable Raman instrument makes the technique practical for trace analysis in clinics, field, and urban settings (29,41).
SERS intensity has been shown to be dependent upon the excitation of the LSPR (29,42–44). Consequently, it is important to control all of the factors affecting the LSPR to maximize signal strength and ensure reproducibility. These factors, which include material, size, shape, interparticle spacing, and dielectric environment, must be chosen carefully to ensure that the incident laser light excites the LSPR. The term enhancement factor is used to describe the magnitude that the SERS effect increases the intensity of the Raman scattering for a given experimental system. The enhancement factor is calculated by dividing the SERS spectral intensity by the normal Raman scattering intensity after normalizing both with respect to collection time, laser power, and number of molecules present in the sampling volume. Previously, a detailed wavelength-scanned SERS study of benzenethiol adsorbed on Ag nanoparticle arrays revealed that the maximum SERS enhancement factor occurs for excitation wavelengths (λex) slightly to the blue of the LSPR maximum wavelength for adsorbate-covered nanoparticle arrays (42). Additionally, similar observations have been reported based upon plasmon-sampled surface-enhanced Raman excitation spectroscopy (29,43). On the basis of these considerations, the most critical aspect of performing a SERS experiment is choice or fabrication of the noble metal substrates.
One of the most robust SERS substrates in use today is the metal film over nanospheres substrates prepared by NSL (Figure 3). The diameter of the nanospheres (D) and the thickness of the metal film (dm) determine the size distribution of the roughness features and, hence, the optical response. AgFON substrates for SERS measurements using 750- or 785-nm laser excitation were optimized by first measuring the dependence of the reflectance LSPR spectral position (λmin) on nanosphere diameter (Figure 4, red round dots). The near-infrared laser excitation was selected because it reduces the native fluorescence background from microorganisms. Because AgFONs are not optically transparent, the reflectivity minimum was used to locate the LSPR maximum. The LSPR measurements were conducted on the AgFON substrates with nanospheres D = 390, 510, 600, and 720 nm. The LSPR λmin shift toward a longer wavelength with the increase in sphere diameter (Figure 4, red round dots). The reflectivity minimum of an AgFON substrate, with D = 600 nm, is ~753 nm and very close to the laser excitation used in SERS. This substrate is expected to show the largest SERS intensity. Further confirmation obtained by comparing bezenethiol Raman signals from these substrates has been summarized in Figure 4 (blue square dots). The normalized SERS intensities at 1003 cm-1 obtained on the four AgFON substrates are plotted as a function of the polystyrene sphere diameters. The plot peaks at D = 600 nm, indicating that the AgFON substrate fabricated using 600-nm spheres is the best substrate for 750-nm laser excitation. The plasmon-scanned SERS technique has revealed a general rule for optimizing SERS intensity when using substrates with narrow LSPR spectra. The largest SERS enhancement factor is achieved when the energy corresponding to the narrow LSPR λmax close to the energy of the excitation wavelength. This finding yields a general rule for maximizing SERS signals for chemical and biological detections. Therefore, this AgFON substrate was selected as optimal for the bacillus spore detection experiments that follow.
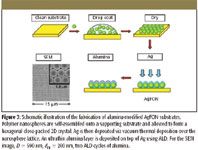
Figure 3
Detection of an Anthrax Biomarker by SERS
The utility of the FON substrate is demonstrated herein as a robust SERS substrate used in the rapid detection of Bacillus subtilis spores, harmless simulants for Bacillus anthracis. A bacillus spore structurally consists of several protective layers and a core cell. CaDPA exists in these protective layers and can be used as the Bacillus and Clostridium spore biomarker because other potentially interfering species lack this particular molecule in such high proportions (45).

Figure 4
CaDPA was extracted from spores by sonicating in 0.02 M HNO3 solution for 10 min. This concentration of the HNO3 solution was selected because of its capability of extraction and benign effect on the AgFON SERS activity. The sonication procedure was performed because no SERS signal of CaDPA was observed from the spore solution before sonication (data not shown). To test the efficiency of this extraction method, a 3.1 × 10-13 M spore suspension (3.7 × 104 spores in 0.2 mL of 0.02 M HNO3) was deposited onto an AgFON substrate (D = 600 nm, dm = 200 nm). The sample was allowed to evaporate for less than one minute. A high signal-to-noise ratio SERS spectrum was obtained in 1 min (Figure 5a). For comparison, a parallel SERS experiment was conducted using 5.0 × 10-4 M CaDPA (Figure 5b). The SERS spectrum of B. subtilis spores is dominated by bands associated with CaDPA, in agreement with the previous Raman studies on bacillus spores (46,47). The SERS spectra in Figure 5, however, display noticeable differences at 1595 cm-1 , which are from the acid form of dipicolinate (48). The peak at 1050 cm-1 in Figure 5a is from the symmetrical stretching vibration of NO3- . Because of its prominence, this peak is used as an internal standard to reduce the sample-to-sample deviations.
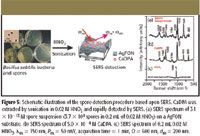
Figure 5
The quantitative relationship between SERS signal intensity and spore concentration is demonstrated in Figure 6a. Each data point represents the average intensity at 1020 cm-1 from three samples with the standard deviation shown by the error bars. At low spore concentrations, the peak intensity increases linearly with concentration (Figure 6a inset). At higher spore concentrations, the response saturates as the adsorption sites on the AgFON substrate become fully occupied. Saturation occurs when the spore concentrations exceed ~2.0 × 10-13 M (2.4 × 104 spores in 0.2 mL of 0.02 M HNO3).
To determine the adsorption capacity of extracted CaDPA on an AgFON, the Langmuir adsorption isotherm was used to fit the data (49,50):
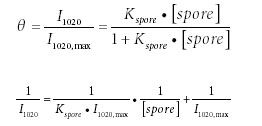
where θ is the coverage of CaDPA on the AgFON; I1020 max is the maximum SERS signal intensity at 1020 cm-1 when all the SERS active sites on AgFON are occupied by CaDPA; [spore] is the concentration of spores (M); and Kspore is the adsorption constant of CaDPA extracted from spores on AgFON (M-1 ). From equation 2, Kspore is calculated from the ratio between the intercept and the slope. Slope and intercept analyses of the linear fit (Figure 6b) lead to the value of the adsorption constant Kspore = 1.7 × 1013 M-1 .
Ultrastable Substrates for SERS: Al2O3 Overlayers Fabricated by Atomic Layer Deposition Yield Improved Anthrax Biomarker Detection
An ideal detection system should run unattended for long periods of time, require infrequent maintenance, and operate at low cost. However, the acceptance of SERS as a general analytical tool has been hindered by the lack of SERS substrate stability. Generally, SERS activity is affected by the oxidation of silver or the aggregation of noble metal colloidal nanostructures (51–53). In this work, we demonstrate a simple strategy for improving the stability of traditional SERS substrates dramatically. An ultrathin alumina layer was coated onto AgFON substrate using atomic layer deposition (ALD) (31). ALD utilizes self-limiting surface reactions to control interfacial thickness and composition with molecular precision (54). Previous quartz crystal microbalance measurements have demonstrated highly uniform layer-by-layer growth of the ALD alumina on Ag nanoparticles with a growth rate of ~1–2 Å per deposition (54). The sub-1-nm thickness is extremely advantageous in preserving sensitivity, because the SERS intensity decays by approximately one order of magnitude for each 2.8 nm of separation between the surface and the scatterer (55). The details of the intensity decay function are determined by the nanostructure of the underlying silver surface.

Figure 6
The use of ALD alumina presents several advantages. First, compared with conventional overlayer materials, the ultrathin alumina layer is extremely stable to oxidation and high temperature (56). This helps to maintain the high stability of SERS activity with minimal decrease in signal. Second, alumina is commonly used as a polar adsorbent in chromatographic separations. The relative affinity between Raman scatterers and alumina-modified AgFON substrates can be predicted based upon their polar interaction, which has been well established in the chromatography literature. Generally, molecules with strong polarity, such as carboxylic acids, have high affinity to alumina (57,58). Therefore, this novel SERS substrate is an ideal candidate for the detection of carboxylic acids due to the strong polar interaction. Third, the scope of analytical applications of SERS has been broadened by modifying noble metal surfaces with an analyte-specific affinity coating (32,59,60). The coatings used range from simple alkanes (59) to complex macrocycles, with the common theme of containing a thiol group to anchor the coating to a noble metal substrate (61). Large partition coefficients on the coating allow analytes to partition closer to the surface (59). However, the high coverage of the thiolate self-assembled monolayers is thermodynamically unstable (62). Thermal desorption (63,64) and photo-oxidation (65–67) of the thiolate molecules result in defects in the coating. In comparison to the previously used thiolate self-assembled monolayers, ALD alumina enjoys greater molecular thickness control, greater physical and chemical stability, more complete surface coverage, less signal attenuation due to distance effects, and predictable affinity. Previously, we have demonstrated that the AgFON substrates modified with two ALD cycles of alumina optimize for the detection of CaDPA, considering the tradeoff between SERS intensity increase due to the greater binding affinity of CaDPA toward alumina compared to silver and the intensity decrease due to the decay of the electromagnetic field around silver nanostructures is demonstrated (68).
The temporal stability of the AgFON substrates modified with two ALD cycles of alumina was studied over a period of 12 months. SERS spectra of 4.7 × 10-14 M spores (3.6 × 103 spores in 0.2 mL of 0.02 M HNO3) were captured on the alumina-modified AgFON substrates of different ages (Figure 7). Figure 7 represents the spore spectrum on a nine-month-old AgFON substrate. The intensity ratios I1020/I1050 were measured to quantitatively compare the substrates of different ages (shown in Figure 7 inset). Both the CaDPA spectral band positions and the intensity patterns remained constant over the course of 12 months, indicating the long-term stability of the AgFON as SERS substrates for potential field sensing applications.

Figure 7
Illustrated in Figure 8, the parallel studies of SERS intensities at 1020 cm-1 versus spore concentrations indicate that the LOD is 1.4 × 103 spores in 0.2 mL of 0.02 M HNO3 and the adsorption constant for CaDPA extracted from spores, Kspore, is 9.0 × 1013 M-1 . In contrast, the adsorption constant was 1.7 × 1013 M-1 for extracted CaDPA on bare AgFON surface and the LOD was 2.6 × 103 spores (laser excitation = 750 nm, laser power = 50 mW, acquisition time = 60 s) (29).

Figure 8
Future Prospects
Alumina-Modified SERS Substrates: The Combination of Separation and SERS Detection
In the analysis of biologically, industrially, and environmentally relevant samples, multiple analytes with similar molecular structures are often in competition for the SERS surface. In that situation, the detection selectivity is improved when the affinity between the target analyte and the sensing platform is optimized. Central to the work reported here is our ability to predict the relative affinity of different molecules on an alumina-modified AgFON substrate. As a proof-of-concept experiment, we examine the competitive adsorption of dipicolinic acid (labeled as 1), diacetylpyridine (labeled as 2), and dimethoxypyridine (labeled as 3) onto an alumina-modified AgFON surface at room temperature (Figure 9a). These pyridine derivative analytes serve as a model system because they provide the unambiguous SERS spectral signatures important to differentiate between the relative adsorption affinities observed during competitive adsorption experiments. On the basis of a TLC experiment (Figure 9b), the relative affinity between each analyte and alumina is determined to be dipicolinic acid > diacetylpyridine > dimethoxypyridine. Therefore, we expect that the SERS spectra of mixed analytes on the alumina-modified AgFON substrates would be dominated by the strongest adsorbate, dipicolinic acid. To verify this hypothesis, a series of SERS measurement were made. We produced the SERS samples by incubating the alumina-modified AgFON substrates in 2 mM analyte in ethanol solutions for 5–6 h. Adsorption of each individual analyte produces a specific SERS signature (Figure 9c). High signal-to-noise SERS spectra were obtained on alumina-modified AgFON substrates even though the adsorbates were not in direct contact with the silver surface. The spectra can be compared with those from equimolar mixtures of pairs of 1 + 2, 1 + 3, and 2 + 3. For mixtures that contain 1, the spectra are dominated by peaks characteristic of 1 at 1458, 1390, 1042, and 1018 cm-1 . Similarly, the spectrum for 2 + 3 combinations is dominated by signatures of 2. From these results, it is clear that 1, dipicolinic acid, has the highest adsorption affinity on alumina-modified AgFON surfaces and 3, dimethoxypyridine, has the lowest, which is consistent with anticipation in light of the TLC results. The high affinity of dipicolinic acid has numerous practical implications. As an example, alumina-modified AgFON surfaces are ideal sensing platforms for calcium dipicolinate, a well-known biomarker for bacillus spores (69), because the increase in affinity improves the ultimate LOD.

Figure 9
The aforementioned experiment demonstrates two very important future trends in SERS. First, the addition of the alumina layer imparts a new chemical functionality to the SERS active surface. We will expand our palette of available ALD layer materials, which will enable greater chemical control over the nature of the selectively adsorbed analytes. Second, the novel SERS substrates reported herein hold enormous promise for combining both TLC and SERS without extra deposition steps of silver colloids or positively charged polymer (70,71). This combination, with the development of miniaturized Raman spectrometers and nanofabrication methods, will allow for effective separation and sensitive identification of components in a mixture analyte, which is an important task for the development of chip-based chemical and biological detection systems.
Tip-Enhanced Raman Spectroscopy
A new technique, tip-enhanced Raman spectroscopy (TERS) as an apertureless optical near-field technique, has great potential to provide material, surface, and crystallographic generality for SERS. The tip of an atomic force microscopy (AFM) or a scanning tunneling microscopy (STM) device provides a locally confined appreciable enhancement of the electromagnetic field of an incoming light wave, thus causing the enhanced Raman scattering from a narrow section of the surface area. In addition, TERS overcomes, in principle, the two obstacles of the standard SERS, namely, the need for nonplanar (rough) surfaces and its restriction to specific adsorbates. Additionally, the Raman enhancement can be probed at any location of the sample.
Multiplexing
LSPR biosensors have been exploited to detect a variety of biotargets, such as DNA, immunoassay, and disease biomarkers. LSPR sensors have been developed based upon patterned nanostructures, metallic films, and single nanoparticles. With the improvement in instrumental resolution, LSPR is promising for the real-time study of surface-binding kinetics. Besides sensing, LSPR can be used to study any process that will lead to a spectral change (for example, pH, humidity, chemical reactions, and small signaling molecules).
There have been some initial efforts to develop multiplexed LSPR sensors. A multiplexed sensing chip has several advantages, including rapid and parallel detection of multiple targets on a single chip, minimized sample-to-sample variation, lower cost per chip, reduced analyte volume, and reduced time for total number of experiments. Yonzon and colleagues have demonstrated a primitive 2 × 1 multiplexed LSPR sensor (14). A (2 × 1) LSPR carbohydrate-binding protein sensing chip consists of a glass substrate with nanoparticle arrays functionalized with galactose and mannose, respectively. To prevent the mixing of the sugar thiol solutions, the nanoparticle array elements were separated by a separator during carbohydrate functionalization. Upon removal of the partition, the LSPR sensor was exposed to ConA. The LSPR λmax of the mannose-functionalized nanoparticles clearly shifted red, indicating the binding of ConA, whereas LSPR λmax of galactose-functionalized nanoparticles did not shift.
Efforts toward engineering a larger array of multiplexed LSPR sensor are in progress. Microfluidic channels currently are being employed to functionalize nanoparticles with various receptors followed by exposure of the protein or proteins of interest to the surface. This type of multiplexed sensor will allow understanding between proteins and their receptors, which will revolutionize the field of cell biology.
Acknowledgments
This work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-03ER15457), the National Science Foundation (EEC-0118025, CHE-0414554, DMR-0520513, BES-0507036), the Air Force Office of Scientific Research MURI program (Grant F49620-02-1-0381), and the National Cancer Institute (1 U54 CA119341-01). The ALD experiments were conducted at Argonne National Laboratory and were supported by the U.S. Department of Energy, BES-Materials Sciences (W-31-109-Eng-38). The authors also would like to thank the Electron Microscopy Center at Argonne National Laboratory, which is supported by the DOE Office of Science (W-31-109-Eng-38).
References
(1) B. Wiley, Y.G. Sun, J.Y. Chen, H. Cang, Z.Y. Li, X.D. Li, and Y.N. Xia, Mrs Bulletin30, 356–361 (2005).
(2) A.J. Mieszawska, R. Jalilian, G.U. Sumanasekera, and F.P. Zamborini,
J. Am. Chem. Soc.127, 10822–10823 (2005).
(3) L.L. Zhao, L. Jensen, and G.C. Schatz,
J. Am. Chem. Soc.128, 2911–2919 (2006).
(4) C.E. Talley, J.B. Jackson, C. Oubre, N.K. Grady, C.W. Hollars, S.M. Lane, T.R. Huser, P. Nordlander, and N.J. Halas, Nano Lett.5, 1569–1574 (2005).
(5) J. Coates, Spectroscopy21(2), 68–74 (2006).
(6) S. Lofas, M. Malmqvist, I. Ronnberg, E. Stenberg, B. Liedberg, and I. Lundstrom, Sens. Actuators B-Chem.5, 79–84 (1991).
(7) M. Malmqvist, Nature361, 186–187 (1993).
(8) A.C. Templeton, J.J. Pietron, R.W. Murray, and P. Mulvaney, J. Phys. Chem. B 104, 564–570 (2000).
(9) T.R. Jensen, M. Duval Malinsky, C.L. Haynes, and R.P. Van Duyne, J. Phys. Chem. B 104, 10549–10556 (2000).
(10) M.A. Schwarz and P.C. Hauser, Lab on a Chip1, 1–6 (2001).
(11) J.M. Brockman, B.P. Nelson, and R.M. Corn, Annu. Rev. Phys. Chem.51, 41–63 (2000).
(12) H.J. Lee, Y.L. Yan, G. Marriott, and R.M. Corn, J. Physiol. (London)563, 61–71 (2005).
(13) M.D. Malinsky, K.L. Kelly, G.C. Schatz, and R.P. Van Duyne, J. Am. Chem. Soc. 123, 1471–1482 (2001).
(14) C.R. Yonzon, E. Jeoung, S.L. Zou, G.C. Schatz, M. Mrksich, and R.P. Van Duyne, J. Am. Chem. Soc.126, 12669–12676 (2004).
(15) D.L. Jeanmaire and R.P. Van Duyne, J. Electroanal. Chem.84, 1–20 (1977).
(16) C.L. Haynes, C.R. Yonzon, X.Y. Zhang, and R.P. Van Duyne, J. Raman Spectrosc.36, 471–484 (2005).
(17) A.J. Haes and R.P. Van Duyne, J. Am. Chem. Soc.124, 10596–10604 (2002).
(18) J.C. Riboh, A.J. Haes, A.D. McFarland, C.R. Yonzon, and R.P. Van Duyne, J. Phys. Chem. B 107, 1772–1780 (2003).
(19) A.J. Haes, L. Chang, W.L. Klein, and R.P. Van Duyne, J. Am. Chem. Soc. 127, 2264–2271 (2005).
(20) A. Dahlin, M. Zach, T. Rindzevicius, M. Kall, D.S. Sutherland, and F. Hook, J. Am. Chem. Soc. 127, 5043–5048 (2005).
(21) C.R. Yonzon, E. Jeoungf, S.L. Zou, G.C. Schatz, M. Mrksich, and R.P. Van Duyne, J. Am. Chem. Soc.126, 12669–12676 (2004).
(22) A.J. Haes, S.L. Zou, J. Zhao, G.C. Schatz, and R.P. Van Duyne, J. Am. Chem. Soc.128, 10905–10914 (2006).
(23) J. Zhao, A. Das, X. Zhang, G.C. Schatz, S.G. Sligar, and R.P. Van Duyne, J. Am. Chem. Soc.128, 11004–11005 (2006).
(24) J.D. Lipscomb and I.C. Gunsalus, Drug Metab. Dispos.1, 1–5 (1973).
(25) I. Schlichting, J. Berendzen, K. Chu, A.M. Stock, S.A. Maves, D.E. Benson, B.M. Sweet, D. Ringe, G.A. Petsko, and S.G. Sligar, Science287, 1615–1622 (2000).
(26) Z.Q. Tian and B. Ren, Annu. Rev. Phys. Chem.55, 197–229 (2004).
(27) K. Kneipp, H. Kneipp, and J. Kneipp, Acc. Chem. Res.39, 443–450 (2006).
(28) M.B. Wabuyele and T. Vo-Dinh, Anal. Chem.77, 7810–7815 (2005).
(29) X. Zhang, M.A. Young, O. Lyandres, and R.P. Van Duyne, J. Am. Chem. Soc. 127, 4484–4489 (2005).
(30) J.K. Daniels, T.P. Caldwell, K.A. Christensen, and G. Chumanov, Anal. Chem.78, 1724–1729 (2006).
(31) X. Zhang, J. Zhao, A.V. Whitney, J.W. Elam and R.P. Van Duyne, J. Am. Chem. Soc.128, 10304–10309 (2006).
(32) C.R. Yonzon, C.L. Haynes, X.Y. Zhang, J.T. Walsh, and R.P. Van Duyne, Anal. Chem.76, 78–85 (2004).
(33) O. Lyandres, N.C. Shah, C.R. Yonzon, J.T. Walsh, M.R. Glucksberg, and R.P. Van Duyne, Anal. Chem.77, 6134–6139 (2005).
(34) D.A. Stuart, C.R. Yonzon, X. Zhang, O. Lyandres, N.C. Shah, M.R. Glucksberg, J.T. Walsh, and R.P. Van Duyne, Anal. Chem.77, 2013 (2005).
(35) K. Kneipp, A.S. Haka, H. Kneipp, K. Badizadegan, N. Yoshizawa, C. Boone, K.E. Shafer-Peltier, J.T. Motz, R.R. Dasari, and M.S. Feld, Appl. Spectrosc. 56, 150–154 (2002).
(36) N. Sundararajan, D.Q. Mao, S. Chan, T.W. Koo, X. Su, L. Sun, J.W. Zhang, K.B. Sung, M. Yamakawa, P.R. Gafken, T. Randolph, D. McLerran, Z.D. Feng, A.A. Berlin, and M.B. Roth, Anal. Chem.78, 3543–3550 (2006).
(37) C.M. Ruan, W. Wang, and B.H. Gu, Anal. Chem.78, 3379–3384 (2006).
(38) D.A. Stuart, K.B. Biggs, and R.P. Van Duyne, Analyst131, 568–572 (2006).
(39) H. Grebel, Z. Iqbal, and A. Lan, Chem. Phys. Lett. 348, 203–208 (2001).
(40) K. Kneipp, H. Kneipp, P. Corio, S.D.M. Brown, K. Shafer, J. Motz, L.T. Perelman, E.B. Hanlon, A. Marucci, G. Dresselhaus, and M.S. Dresselhaus, Phys. Rev. Lett.84, 3470–3473 (2000).
(41) K. Chen, M.E. Martin, and T. Vo-Dinh, Proc. SPIE-Int. Soc. Opt. Eng. 5993, 599307/599301-599307/599308 (2005).
(42) A.D. McFarland, M.A. Young, J.A. Dieringer, and R.P. Van Duyne, J. Phys. Chem. B 109, 11279–11285 (2005).
(43) C.L. Haynes and R.P. Van Duyne, J. Phys. Chem. B 107, 7426–7433 (2003).
(44) J.B. Jackson and N.J. Halas, Proc. Natl. Acad. Sci. U.S.A.101, 17930–17935 (2004).
(45) R. Goodacre, B. Shann, R.J. Gilbert, E.M. Timmins, A.C. McGovern, B.K. Alsberg, D.B. Kell, and N.A. Logan, Anal. Chem.72, 119–127 (2000).
(46) S. Farquharson, A.D. Gift, P. Maksymiuk, and F.E. Inscore, Appl. Spectrosc.58, 351–354 (2004).
(47) S. Farquharson, L. Grigely, V. Khitrov, W. Smith, J.F. Sperry, and G. Fenerty,
J. Raman Spectrosc.35, 82–86 (2004).
(48) P. Carmona, Spectrochim. Acta, Part A36A, 705–712 (1980).
(49) L.S. Jung and C.T. Campbell, J. Phys. Chem. B 104, 11168–11178 (2000).
(50) L.S. Jung and C.T. Campbell, Phys. Rev. Lett.84, 5164–5167 (2000).
(51) K.U. Von Raben, R.K. Chang, B.L. Laube, and P.W. Barber, J. Phys. Chem.88, 5290–5296 (1984).
(52) J.H. Fang, Y.X. Huang, X. Li, and X.M. Dou, J. Raman Spectrosc.35, 914–920 (2004).
(53) D. Fornasiero and F. Grieser, J. Chem. Phys.87, 3213–3217 (1987).
(54) A.V. Whitney, J.W. Elam, S.L. Zou, A.V. Zinovev, P.C. Stair, G.C. Schatz, and R.P. Van Duyne, J. Phys. Chem. B 109, 20522–20528 (2005).
(55) J.A. Dieringer, A.D. McFarland, N.C. Shah, D.A. Stuart, A.V. Whitney, C.R. Yonzon, M.A. Young, X.Y. Zhang, and R.P. Van Duyne, Faraday Discuss.132, 9–26 (2006).
(56) F. King, in Aluminum and Its Alloys, E.G. West, ed. (Ellis Horwood, New York, 1987), p. 313.
(57) D.L. Allara and R.G. Nuzzo, Langmuir1, 45–52 (1985).
(58) D.L. Allara and R.G. Nuzzo, Langmuir 1, 52–66 (1985).
(59) K. Carron, L. Peitersen, and M. Lewis, Environ. Sci. Technol.26, 1950–1954 (1992).
(60) J.D. Driskell, K.M. Kwarta, R.J. Lipert, M.D. Porter, J.D. Neill, and J.F. Ridpath, Anal. Chem.77, 6147–6154 (2005).
(61) T.O. Deschaines and K.T. Carron, Appl. Spectrosc.51, 1355–1359 (1997).
(62) J.C. Love, L.A. Estroff, J.K. Kriebel, R.G. Nuzzo, and G.M. Whitesides, Chem. Rev.105, 1103–1169 (2005).
(63) T. Ishida, M. Hara, I. Kojima, S. Tsuneda, N. Nishida, H. Sasabe, and W. Knoll, Langmuir14, 2092–2096 (1998).
(64) Z.S. Zhang, O.M. Wilson, M.Y. Efremov, E.A. Olson, P.V. Braun, W. Senaratne, C.K. Ober, M. Zhang, and L.H. Allen, Appl. Phys. Lett.84, 5198–5200 (2004).
(65) M. Lewis, M. Tarlov, and K. Carron,
J. Am. Chem. Soc. 117, 9574–9575 (1995).
(66) M.H. Schoenfisch and J.E. Pemberton, J. Am. Chem. Soc.120, 4502–4513 (1998).
(67) Y.M. Zhang, R.H. Terrill, T.A. Tanzer, and P.W. Bohn, J. Am. Chem. Soc. 120, 2654–2655 (1998).
(68) X. Zhang, J. Zhao, A.V. Whitney, J.W. Elam, and R.P. V. Duyne, J. Am. Chem. Soc.128, 10304–10309 (2006).
(69) G.F. Bailey, S. Karp, and L.E. Sacks,
J. Bacteriol.89, 984–987 (1965).
(70) J.M.L. Sequaris and E. Koglin, Anal. Chem.59, 525–527 (1987).
(71) R.M. Seifar, M.A.F. Altelaar, R.J. Dijkstra, F. Ariese, U.A.T. Brinkman, and C. Gooijer, Anal. Chem.72, 5718–5724 (2000).
(72) J.W. Elam, M.D. Groner, and S.M. George, Rev. Sci. Instrum.73, 2981–2987 (2002).
(73) M.D. Groner, F.H. Fabreguette, J.W. Elam, and S.M. George, Chem. Mater. 16, 639–645 (2004). n
Pittcon 2025: Keynote Coulter Lecture Highlights Work in Regenerative Engineering
March 3rd 2025Yesterday, at 5:00 pm in Ballroom East, the Wallace H. Coulter Lecture took place, and it was delivered by Cato T. Laurencin, MD, PhD, who is well-known as a scientist and entrepreneur with an extensive career in regenerative engineering. His lecture highlighted the work he and his team has done in this space.
Advancing Zebrafish Research: FT-IR Imaging Sheds Light on Tissue Preservation in Zebrafish
February 5th 2025Researchers at the University of Lublin and the Medical University of Lublin have demonstrated the first application of FT-IR imaging in zebrafish larvae, revealing that frozen samples better preserve tissue structure than chemical fixation.
