Using LC–oa-TOF MSE with a Multivariate Statistical Sample Statistical Sample Profiling Strategy to Distinguish Chinese Red Ginseng from Korean Red Ginseng
Special Issues
Both Chinese ginseng and Korean ginseng are similar plant species and undergo similar handling procedures when harvested and processed for sale. Despite their similarities, Korean ginseng commands a higher price than Chinese ginseng on the open market and is believed to produce different clinical effects than Chinese ginseng. Chinese researchers are now employing new techniques on the two varieties of ginseng to understand their chemical differences. HPLC/UV-based strategies for distinguishing the two types of ginseng have proven to be mostly ineffective due to lack of resolution. Using UltraPerformance liquid chromatography/orthogonal acceleration (oa)–TOF mass spectrometry and exact mass measurement, the authors developed a high-resolution method using multivariate statistical analysis for separating and identifying differences between Chinese ginseng and Korean ginseng at the molecular level.
Ginseng is a plant in the Panax genus found in the Northern Hemisphere and in East Asian countries. It is commonly used medicinally by East Asian cultures. The root of ginseng is promoted as an adaptogen (a product that increases the body's resistance to stress), an aphrodisiac, and a stimulant and is often an ingredient in functional foods such as energy drinks (1). As a traditional Chinese medicine, once harvested, the root of ginseng is steamed and dried into a product that is called red ginseng. Presently, there are two primary types of red ginseng in the traditional Chinese medicine formulary: one is Chinese red ginseng and the other is Korean red ginseng.
For many years, Korean red ginseng has been perceived as clinically more effective than Chinese red ginseng. It is not surprising then that Korean red ginseng commands a significantly higher price than Chinese red ginseng in retail stores. Korean red ginseng often is regarded as a luxury product. By all outward appearances, Korean red ginseng and Chinese red ginseng smell, feel, look, and taste very similar. For this reason, it is not uncommon for counterfeit merchandisers to sell Chinese red ginseng as Korean red ginseng.
The main bioactive constituents of ginseng are considered to be triterpene saponins, generally referred to as ginsenosides (2). It has been noted in the literature that clear differences exist in ginsenoside composition and relative abundance among Panax roots harvested from different geographical locations (3–5). However, Korean red ginseng and Chinese red ginseng share the same geographic origin, and apart from undergoing different processing procedures, no significant differences in their ginsenoside content have been identified (6,7). Presently, there are no routine analytical methods available to effectively distinguish the chemical contents of the two products.
For these reasons, we systematically compared the chemical content of Korean red ginseng and Chinese red ginseng with a sample profiling strategy combining UltraPerformance LC (Waters Corporation, Milford, Massachusetts), orthogonal acceleration time-of-flight mass spectrometry (oa-TOF-MS), and multivariate statistical analysis (MVA) as the analytical tools (8). Our hope is that this will open a door of exploration that allows us to understand the subtle differences between some common natural products like Korean red ginseng and Chinese red ginseng.
Experimental
Ginseng Samples
For this study, a total of 20 red ginseng samples were collected by the Beijing Institute for Drug Control. Among them, four were Chinese red ginseng purchased from four separate TCM drug stores in China, and they are labeled as CH1B, CH2S, CH3T, and CH4Y. The other 16 samples were Korean red ginseng that belonged to three different categories: Tian (TX, T10, T20, T30, T30, T40, T50, T60, and T80); Di (D30 and D40); and Ren (R20, R30, R40, R40s, R50).
The label codes for the Korean red ginseng reveal the sizes of the ginseng roots. For example, T10 refers to Korean red ginseng from the Tian group, and contains 10 pieces of root per 500 g, indicating that the root in this sample is fairly large. On the other hand, T80 refers to Korean red ginseng from the Tian group that contains 80 pieces per 500 g, indicating that the root in this sample is fairly small. A discussion about root size and its impact on study results appears later in this article. To help better understand the samples, Figure 1 shows the pictures of a Korean red ginseng sample and a Chinese red ginseng sample.

Figure 1: Pictures of Korean red ginseng and Chinese red ginseng.
Sample Preparation
Crude red ginseng root was refluxed with 70% methanol. The extraction solution was concentrated into residue, which was then dissolved in water. The water solution was loaded onto an Oasis HLB solid-phase extraction (SPE) cartridge (Waters Corporation, Milford, Massachusetts). It was washed with 20% methanol first, then with 70% methanol, collected, and evaporated to dryness. The residue was reconstituted with 1 mL of methanol and injected into the LC system after being filtered through a 0.45-μm PTFE membrane.
Reagents
The Fisher Optima grade acetonitrile, methanol, and isopropanol were obtained from ThermoFisher Co. (Waltham, Massachusetts). Formic acid and leucine enkephaline were purchased from Sigma Aldrich (St. Louis, Missouri). Deionized water was obtained in our laboratory via a Milli-Q water purification system (Millipore Corporation, Bedford, Massachusetts).
Liquid Chromatography Conditions
The LC separation was performed on an ACQUITY UPLC system (Waters Corporation, Milford, Massachusetts) with an ACQUITY UPLC HSS T3 column (100 mm × 2.1 mm, 1.7 μm). The column temperature was set at 45 °C. The flow rate was kept at 600 μL/min. Mobile phase A was water with 0.1% formic acid, and mobile phase B was acetonitrile. The initial gradient condition was 95% A, followed by a 1-min linear gradient to 85% A, then to 35% A over 24 min, stepped to 0% A and held for 3 min before being stepped back to 95% A for a 3-min equilibration. The total run time was 30 min, and the injection volume was 2 μL.
Mass Spectrometry Conditions
The MS detection was performed on a Synapt MS System (Waters Corporation). The data acquisition mode was MSE. The ionization mode was negative electrospray (ESI–). The source temperature was set at 120 °C, and the desolvation temperature was set at 450 °C with desolvation gas flow set at 900 L/h. The lock mass compound used was leucine enkephaline. The capillary voltage was set at 3 kV. The cone voltage was set at 40 V. The collision energies were set as 5 eV (trap) and 4 eV (transfer) for low-energy scan, and 40–80 eV ramp (trap) and 10 eV for high-energy scan.
The LC–MS data acquisition was controlled by MassLynx 4.1 Mass Spectrometry Software (Waters Corporation).
Data Processing
Post-acquisition data processing including the MVA was performed by MarkerLynx XS, which is an "application manager" for MassLynx software. The structural elucidation was performed by the MassFragment tool provided by MassLynx.
Results and Discussion
LC–TOF MSE Multivariate Statistical Small Molecule Sample-Profiling Strategy
The most critical part of any analytical protocol for profiling of a complex sample is the ingredient separation. The quality of the final result of an analytical protocol is directly related to the quality of the initial chromatographic separation. Ingredients must be separated physically from one another in order for them to be detected properly and for the detailed sample profile to be obtained. The better the separation, the easier it is to differentiate closely related samples.
LC has been the dominant analytical separation technique in laboratories for decades. With the introduction of columns packed with sub-2-μm particles in the first half of this decade, and specially designed instruments capable of withstanding back pressures up to 15,000 psi, separations are now routinely performed with much greater speed, resolution, and sensitivity (9).
Resolution is a key consideration when developing an analytical protocol for complex sample profiling. While speed is important, it is meaningful only when sufficient separation resolution is obtained. Samples of a natural origin like ginseng typically contain hundreds of ingredients. Sufficient separation of these ingredients requires sufficient peak capacity during the sample analysis. For this reason, the protocol for this project utilized a 30-min run time from injection to injection.
When coupled with LC, MS is a widely accepted form of detection, especially for sample-profiling purposes, due to its selectivity and sensitivity. When profiling complex samples like Korean red ginseng and Chinese red ginseng, LC combined with UV detection has its limitations. For example, Figure 2 shows an HPLC–UV trace previously obtained for a red ginseng in which only limited information is revealed with just a few peaks detected.

Figure 2: Previous HPLCâUV result obtained from a red ginseng sample.
Of various mass spectrometers available to choose from, we chose an orthogonal acceleration time-of-flight detector for our sample profiling work. A TOF mass spectrometer offers the required sensitivity and is capable of making exact mass measurements for compounds detected, allowing elemental composition to be obtained for sample analytes. In addition, by coupling with a quadrupole and a collision cell in sequence in front of the flight tube, the instrument filters out unwanted ions so that only ions with masses selected by the quadrupole will enter the flight tube for detection, increasing selectivity. This instrument setting also allows MS-MS data to be collected for structural elucidations when there is a need. Figure 3 shows our entire analytical workflow for small molecule sample profiling.
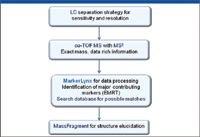
Figure 3: Small molecule sample profiling workflow for LCâoa-TOF-MSE with multivariate statistical analysis.
In this workflow, we utilized the MSE data acquisition strategy shown in Figure 4 (10,11). This strategy leverages the advantages of UHPLC for high resolution, high sensitivity, and high-speed separation, as well as the TOF exact mass measurement capability. This data acquisition strategy allows the mass spectrometer to obtain two distinct but parallel data sets, one at low collision energy (CE) and the other at high CE. The resulting raw data file contains two distinct chromatograms; one contains data with the intact mass-to-charge ratios (m/z) while the other contains fragment ion information. From a single LC injection, the MS full scan and fragment ion information can be obtained simultaneously.
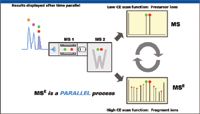
Figure 4: oa-TOF MSE data acquisition strategy.
Figure 5 shows the base peak ion chromatograms (BPI) obtained for the red ginseng extracts using the data-acquisition strategy. Figure 5a is the BPI of the Chinese red ginseng and Figure 5b is the BPI of the Korean red ginseng. The BPIs shown here were obtained from the low CE scan because these data are used for the initial screening of the key markers. The high CE scan data are used for structural elucidation at a later stage when a chemical structure of a specific marker is identified.
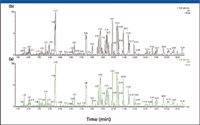
Figure 5: Base peak ion chromatograms of Chinese ginseng (a) versus Korean ginseng (b).
Because of the complexity of the sample (Figure 5), it is difficult to distinguish between the Korean red ginseng and the Chinese red ginseng by visual observation the two chromatograms. This is also an indication that the major ingredients of the two samples may be very similar. To identify the differences between the samples chemically, an effective approach is the MVA. MVA has been used widely in the metabonomics field in recent years for samples of extreme complexity (12).
One of the prerequisites for applying MVA to LC–MS data is that the data must be in a 2-D format. Each data point on the chromatographic trace shown in Figure 5 is actually a 3-D data point; it represents retention time, m/z, and intensity. This 3-D LC–MS dataset must be reduced into a 2-D matrix before it can be analyzed by MVA. This is accomplished automatically by the MarkerLynx XS software.
The software converts each data point into an exact mass retention time (EMRT) pair and tabulates the results into a 2-D matrix. Figure 6 shows the 2-D tabulated results obtained for the Korean red ginseng and the Chinese red ginseng comparison experiment. When using MVA for sample analysis, it is crucial that each sample is injected a minimum of three times to ensure that the data analysis is statistically valid. For our work, each red ginseng sample was injected six times.
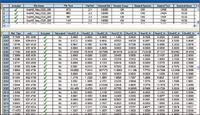
Figure 6: MarkerLynx XS browser window. Top window shows the list of sample injections. The bottom window shows the exact mass retention time table.
After the EMRT 2-D matrix is obtained, the MVA interface will be launched with all EMRT information automatically imported so that the extended statistics module principal component analysis (PCA) can be carried out. PCA analysis is a commonly used, unsupervised statistical tool to observe the grouping patterns of samples (13). From the PCA plot, any two groups of samples can be paired for an orthogonal partial least squares-data analysis (OPLS-DA) (14). The resulting scatter plot (S-plot) from the OPLS-DA analysis can clearly display significant markers that differentiate the sample groups.
Comparison of the Korean Red Ginseng Versus the Chinese Red Ginseng
Figure 7 shows the PCA scores plot obtained from the low CE scan of the LC–oa-TOF-MSE data analysis. Each of the red ginseng samples is clearly labeled in the figure. The first observation here is that there is not a clear distinction between the Chinese red ginseng samples and the Korean red ginseng samples. It appears that some Chinese red ginsengs are chemically more similar to certain Korean red ginsengs. For example, the Chinese red ginsengs CH2H and CH4Y are more closely grouped with the Korean red ginsengs KRR20, KRT20, KRT10, KRT30, and KRD 40S; while the Chinese red ginseng CH3T is more closely grouped with the KRD30, KRT30, KRT40, KRT50, KRT60, KRT80, KRTX, KRR30, KRR40, KRR40S, and KRR50. And the Chinese red ginseng CH1B is distinctively different from all other samples and grouped by itself.

Figure 7: PCA scores plot of the Korean red ginseng verdud the Chinese red ginseng.
This result may provide a possible explanation as to why clear differences have not been reported so far between the Korean red ginseng and the Chinese red ginseng. The experimental results might have been affected by the type of samples selected for the study and the processing method of the samples.
The second observation is that the size of the roots from which samples are taken does affect ingredient compositions, as shown in Figure 7. If a line were to be drawn across the PCA plot (shown in Figure 7), all groups above the line are the ginseng samples taken from larger-sized roots (KRT 10 to KRD 40S), while the samples grouped below the line are from the smaller-sized ginseng roots (KRT 30 to KRT 80). In the ginseng market, the size of the ginseng root is also a price differentiator. This observation from the PCA plot indicates that this practice may not be entirely scientifically unfounded.
A useful approach for MVA analysis is to focus the analysis on two groups and use a supervised statistical model referred to as orthogonal projection to latent structural-discriminate analysis (OPLS-DA). An orthogonalized scores plot is then obtained, clearly displaying the differences between the two sample groups. From the loading plot, in S-plot format, the markers contributing to the differences between the two sample groups can be identified easily (8).
In our analysis, even though the initial goal was to compare Chinese red ginseng to Korean red ginseng, the PCA analysis result clearly indicated that we effectively have a multiple grouping pattern of our sample set. As a result, the OPLS-DA analysis can be performed for many different grouping pairs. For example, all Chinese red ginseng versus all Korean red ginseng; the CH1B Chinese red ginseng versus all (all other 19 samples as one group); the CH1B Chinese red ginseng versus the other three Chinese red ginseng (the other three Chinese red ginseng as one group); and all small Korean red ginseng versus all large Korean red ginseng according to the grouping pattern shown on the PCA plot.
The general workflow for using OPLS-DA to compare two groups of samples and obtain major markers should be consistently the same regardless of which groups of samples were to be compared. For demonstration purposes, we chose one pair of groups to compare. As shown in Figure 7, the two groups — CH1B versus the KRD 30 — are the farthest apart on the PCA scores plot. The OPLS-DA analysis was performed on these two groups and the OPLS-DA scores plot is shown in Figure 8. From the scores plot, the difference between the two sample groups is displayed along the x-axis, and the injection-to-injection differences within the same sample group are displayed along the y-axis.

Figure 8: PCA scores plot from OPLS-DA analysis for CH1B versus KRD30.
Figure 9 shows the loading plot (S-plot) of the OPLS-DA result. In the S-plot, each point represents an EMRT pair (a marker). The x-axis shows the variable contributions. The further away a data point is from the 0 value, the more it contributes to sample variance. The y-axis shows the sample correlations within the same sample group. The further away an EMRT pair is from the value 0, the better is its correlation from injection to injection. As a result, the EMRT pairs on both ends of the S-shaped curve represent the leading contributing ions from each sample group. For example, in Figure 9, the EMRT pairs nearest the upper right corner of the S-plot are the leading contributing markers from the Chinese red ginseng CH1B group; the EMRT pairs nearest the lower left corner of the S-plot are the leading contributing markers from the Korean red ginseng KRD30 group.

Figure 9: Scatter plot (S-plot) from OPLS-DA analysis for CH1B versus KRD30.
The leading contributing EMRT pairs can be captured selectively so that a list of top contributing markers from each sample group can be generated and saved as a text file. This text file can be imported into a results table for an elemental composition calculation and database search. Table I shows the combined list of the top eight leading EMRT pairs obtained from the S-plot for both sample groups. The markers in red are highest in concentration in the Chinese red ginseng CH1B group, and the markers in blue are highest in concentration in the Korean red ginseng KRD30 group. This table also can be displayed as a bar chart, as shown in Figure 10.

Figure 10: Bar chart showing the major marker changes.
Table I also shows the factor of change for each of the markers identified. It is worth noting that the markers that showed highest change factors are not necessarily those with highest peak intensities. For example, the marker of highest intensity among the markers in Table I is 4.54_845.4920; however, its content reduction from CH1B to KRD 30 is only 1.5, meaning that, relatively speaking, the changing factor of this marker from one group to another is not as significant as some of the markers of lower intensities. For example, the marker 3.93_931.5280 appeared to be the weakest in intensity on the top half of the list; however, its changing factor from one group to another is 8.9. As a result, the most intense markers tend to stretch out more toward the end of the S-plot, but they might not be the ones that show the highest factor of change. For demonstration purposes, we are focusing our discussion on those points taken from both ends of the S-plot. In reality, when analyzing data for any project, the points with smaller intensity values along the x-axis but showing good injection-to-injection correlations are worth spending time to investigate as well.
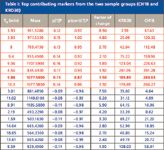
Table I: Top contributing markers from the two sample groups (CH1B and KRD30)
The next step after obtaining the marker list is to perform an elemental composition calculation for the targeting markers. The elemental composition obtained can be used to search against an existing database. For example, the marker 9.88_1077.5800 matched elemental composition of C53H89O22. Therefore, the molecular formula of the marker in question is C53H90O22 because our experiment was conducted under negative electrospray conditions. This elemental composition is then used to search against a database: www.chemspider.com (available in public domain). Figure 11 shows the screen shot of the search result for this marker: ginsenoside Rb2.
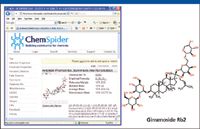
Figure 11: ChemSpider database search result for m/z 1077.5800.
Once the identity of a marker is tentatively identified, its fragment ion can be obtained easily by going back to the raw data file to investigate the high CE scan of the sample. The fragment ions can be used for structural elucidation using the MassFragment tool available with MarkerLynx for added confidence in marker identification and for guiding the direction of the project. As an example, Figure 12 shows the fragment ion structure obtained automatically by MassFragment for the ginsenoside Rb2.

Figure 12: Structural elucidation of the marker 1077.5800 fragment ions by MassFragment software.
As previously discussed, the change factor of the ginsenoside Rb2 is less than 2, which is not as significant. This might provide another explanation for the fact that previous studies could not identify the differentiating ingredients from the Chinese red ginseng and the Korean red ginseng. For example, a marker 3.01_m/z 861.4856 shows change factor of 7.5 from CH1B to KRD30; however, its intensity is much lower than that of the ginsenoside Rb2. Figure 13 shows the screen shot of the database search result indicating that it is one of the less common ginsenosides. This shows that even though there are only small-scale changes observed among the major ginsenosides from one ginseng sample to another, changes in concentration of some less commonly discussed ginsenosides can be more significant, but they may not have been identified in previous comparative studies.
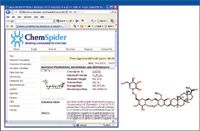
Figure 13: Chemspider database search result for m/z 861.4856.
Conclusion
The generic LC–oa-TOF-MSE sample profiling strategy allows multiple groups of complex samples to be studied by using the multivariate statistical analysis approach. The combination of the high-resolution LC separation and high-resolution MS detection along with the multivariate statistical analysis allowed details of the samples to be profiled so that important markers of variance even at low concentration levels can be measured.
As a result, this is the first time that the differences between Korean red ginseng and Chinese red ginseng have been observed systematically and at the chemistry level.
Kate Yu, Jose Castro-Perez, John Shockcor, and Brian Murphy are with Waters Corporation, Milford, Massachusetts. Hongzhu Guo, Mintong Xin, Xintong Fu, and Yougen Chen are with Beijing Institute for Drug Control, Beijing, P.R. China.
References
(1) D. Winston and S. Maime,.Adaptogens: Herbs for Strength, Stamina, and Stress Relief (Healing Arts Press, 2007).
(2) S. Zhu, K. Zou, S. Cai, M.R. Meselhy, and K. Komatsu, Chem. Pharm. Bull. 52(8), 995–998 (2004).
(3) O. Tanaka, Pure Appl. Chem. 62, 1281–1284 (1990).
(4) T. Morita, O. Tanaka, and H. Kohda, Chem. Pharm. Bull. 33, 3852–3858 (1985).
(5) O. Tanaka, E.C. Han, Y. Yamaguchi, H. Matsuura, T. Murakami, T. Taniyama, and M. Yoshikawa, Chem. Pharm. Bull. 48, 889–892 (2000).
(6) G. Wang, J. Tian, L. Nie, X. Liang, and R. Ling, Chin. Traditional and Herbal Drugs 39, 277–280 (2008).
(7) C. Zhang, Y. Zheng, L. Sun, X. Li, and Y. Ren, Renshen Yanjiu 13(2), 9–11 (2001).
(8) K. Yu, J. Castro-Perez, and J. Shockcor, Waters Application Note 720002541 (2008).
(9) Waters White paper, Ultra Performance LC UPLC: New Boundaries for the Chromatography Laboratory, 720000819 EN (May 2004).
(10) K. Bateman, J. Castro-Perez, M. Wrona, J. Shockcor, K. Yu, R. Oballa, and D. Nicoll-Griffith, Rapid Commun. Mass Spectrom. 21, 1485–1496 (2007).
(11) P. Tiller, S. Yu, J. Castro-Perez, K. Fillgrove, and T. Baillie, Rapid Commun. Mass Spectrom. 22, 1053–1061 (2008).
(12) I. Wilson, J. Nicholson, J. Castro-Perez, J. Granger, K. Johnson, B. Smith, and R. Plumb, J. Proteome Res. 4, 591–598 (2005).
(13) I.T. Jolliffe, Principal Component Analysis (second edition) (Springer publishing, 2002).
(14) L. Eriksson, E. Johansson, N. Kettaneh-World, J. Trygg, C. Wikstrom, and S. World, Multi- and Megavariate Data Analysis Part II: Advanced Applications and Method Extensions, Second revised and enlarged edition (Umetrics Inc., 2006).

LIBS Illuminates the Hidden Health Risks of Indoor Welding and Soldering
April 23rd 2025A new dual-spectroscopy approach reveals real-time pollution threats in indoor workspaces. Chinese researchers have pioneered the use of laser-induced breakdown spectroscopy (LIBS) and aerosol mass spectrometry to uncover and monitor harmful heavy metal and dust emissions from soldering and welding in real-time. These complementary tools offer a fast, accurate means to evaluate air quality threats in industrial and indoor environments—where people spend most of their time.
NIR Spectroscopy Explored as Sustainable Approach to Detecting Bovine Mastitis
April 23rd 2025A new study published in Applied Food Research demonstrates that near-infrared spectroscopy (NIRS) can effectively detect subclinical bovine mastitis in milk, offering a fast, non-invasive method to guide targeted antibiotic treatment and support sustainable dairy practices.
Smarter Sensors, Cleaner Earth Using AI and IoT for Pollution Monitoring
April 22nd 2025A global research team has detailed how smart sensors, artificial intelligence (AI), machine learning, and Internet of Things (IoT) technologies are transforming the detection and management of environmental pollutants. Their comprehensive review highlights how spectroscopy and sensor networks are now key tools in real-time pollution tracking.