Creating a High-Throughput LC–MS-MS System Using Common Components
Special Issues
How to create a liquid chromatography–tandem mass spectrometry (LC–MS-MS) system using mass spectrometers, a high performance liquid chromatography (HPLC) binary pump system, and an autosampler
A liquid chromatography–two-dimensional mass spectrometry (LC–MS-MS) system that achieves a 72-s injection-to-injection cycle time was created by making several small modifications to common components. The system was used to quantify a wide range of chemical compounds in various absorption, distribution, metabolism, and excretion (ADME) assays with an analytical success rate >90%. Method considerations included the amount of carryover when reducing the number of needle washes, potential loss of signal intensity with increased flow rate, and ideal column pore size to perform runs consisting of more than 2000 injections. These modifications enable almost any laboratory to operate in a high-throughput manner without having to purchase state-of-the-art equipment. In addition, replacing the mass spectrometer with a more sensitive system opens up the possibility of pooling up to six samples per injection, achieving a net injection-to-analyte time of 12 s, which is comparable to the most advanced LC–MS-MS systems currently on the market.
The desire to provide new treatments for unmet diseases or conditions, along with the opportunity to provide improvements in safety and efficacy for existing therapies, highlight the urgency to bring new drug entities (NDEs) to market. Research and development costs to provide these solutions have risen to an estimated $1.8 billion per NDE (1). These needs and costs highlight the importance of bringing a product to market quickly to provide patient benefit as early as possible.
Current efforts to speed up the drug discovery process have included a dramatic increase in chemical synthesis over the past decades through the rise of combinatorial chemistry and the outsourcing of chemical synthesis (2). It is important to note that with this increase in chemistry production comes the demand to provide supporting information critical to the development of a safe, effective, and marketable drug. Supporting information includes measurements of potency, metabolism, elimination pathways, and toxicity, to name just a few. These data are used to promote candidates with acceptable properties to the next stage in the discovery funnel and to drive the study of structure-activity-relationships (SAR). The faster these data can be obtained, the more impact they will have on selecting the right drug candidates earlier in the process, therefore reducing the overall time from discovery to market. Many of the measurements listed above require the use of liquid chromatography–mass spectrometry (LC–MS) for the detection of the drug candidates or other assay endpoints. Therefore, having access to LC–MS instrumentation that can rapidly screen compounds is a distinct advantage, because so many of the assays depend on it for their data endpoints.
Currently, there are several "out of the box" platforms available to increase MS sample analysis throughput. Although such systems are convenient and powerful, their price can exceed several hundreds of thousands of dollars. Complicating the matter further is the fact that many research budgets are either shrinking or remaining stagnant (3,4). Scientists are now being asked to do more with less and to make do with the instrumentation that is currently in their laboratories. This often means working with instrumentation that is not state-of-the-art or purchasing readily available used equipment from auction sites such as DoveBid or Labequip. At the time this article was written, there were three AB Sciex API-3000 mass spectrometers, 11 Agilent 1100 high performance liquid chromatography (HPLC) binary pump systems, and seven CTC Analytics HTS autosamplers available for purchase on DoveBid. This article demonstrates how to create an LC–MS-MS system using only these three common components that approaches a 1-min injection–injection cycle time, without the use of column swapping, multiple LC systems, or aftermarket scheduling software.
Experimental
Instrumentation
The system used in these studies comprised an AB Sciex API-3000 mass spectrometer with a turbo ion spray source from Applied Biosystems (Foster City, California), an Agilent 1100 Series binary pump with a vacuum degasser (Santa Clara, California), a CTC Analytics model HTS-PAL autosampler from LEAP Technologies (Carrboro, North Carolina), and a Valco Cheminert two-position, 10-port switching valve from VICI (Houston, Texas). Fortis Pace C18 columns (30 mm × 2.1 mm) were obtained from Resolution Systems (Holland, Michigan) and were used for all tests unless otherwise stated. Mobile phase A consisted of 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. All solvents were HPLC grade from Sigma-Aldrich (St. Louis, Missouri). A test solution of carbutamide, carbamazepine, imipramine, and verapamil in 50:50 acetonitrile–water was used throughout the process to gauge relative retention times. The compounds were purchased from Sigma-Aldrich.
Initial Method
A baseline for comparative purposes was established using a commonly used gradient and flow rate. The initial method details are as follows: mobile phase A was held at 95% for the first 0.3 min, then decreased to 5% over the next 0.5 min, and held at that level for 0.7 min. Mobile phase A was then increased over the next 0.1 min back to 95% and held for an additional 1.4 min, yielding a 3.0-min method. The flow rate was constant at 500 µL/min for the duration of the method. The column was a Fortis C18 (30 mm × 2.1 mm, 5-µm dp). Tuning of the compounds to obtain precursor and product ion, collision energy, and declustering potential information was performed beforehand and used to generate the multiple reaction monitoring (MRM) methods. All compounds were ionized in positive mode. Using these standard conditions, the analytes were eluted between 2.3 and 2.6 min (Figure 1a) with an injection-to-injection time of 3.4 min after Analyst software "think time" and autosampler arm movement were included.
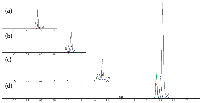
Figure 1: Comparison of retention times by method. Blue = carbutamide, red = imipramine, green = verapamil, grey = carbmazepine. Shown are (a) a 3-min method at a 500-µL/min flow rate with a 400-µL delay volume representing the mixing chamber; (b) a 2-min method at 500 µL/min with the mixing chamber removed; (c) a 1.5-min method at 500 µL/min with a 0.2-min gradient; and (d) a 0.95-min method at 1000 µL/min with a 0.2-min gradient.
Method Changes — HPLC
The first modification investigated was a reduction of the system dead volume. We began with the biggest source of dead volume in this system: the HPLC instrument. Agilent has previously documented how to reduce the dead volumes of its 1100 and 1200 series HPLC systems (5). The detailed procedure involves bypassing both the mixing chamber and dampener. For the work done in these studies, only the mixing chamber was bypassed because the dampener is required to read and report the pressure on the 1100 series HPLC system. In the case of the 1200 series HPLC system, the dampener is not used to measure the system pressure and can therefore be bypassed as an additional option for reducing delay volume. This single adjustment reduced the retention time, as compared to the baseline method, by almost a full minute, with the analytes eluted between 1.4 and 1.7 min (Figure 1b). This would allow an analyst to end the method at 2.0 min, yielding a method capable of 2.4-min injection-to-injection times. It should be noted that to recreate the 400-µL mixing chamber volume on the HPLC system of the initial method, a piece of wide-bore tubing of the same volume was used because our mixing chambers were previously discarded and unavailable for use in testing.
Peaks were being eluted around 1.5 min into the method, so the next alteration was to the gradient profile. We wanted to take advantage of the software "think time" between when one injection ends and the next one starts by returning to original conditions at the very end of the method, allowing the system to re-equilibrate during that "think time." Various gradient profiles were tested before settling on the following: Mobile phase A was held at 95% for the first 0.05 min, decreased to 2% over the next 0.1 min, where it was held for 0.55 min, then increased back to 95% over the next 0.05 min for an additional 0.75 min, resulting in a 1.5-min method. This adjustment reduced the retention times of the analytes to 0.9 and 1.2 min (Figure 1c), and yields a method capable of a 1.9-min injection-to-injection time.
Method Changes — Mass Spectrometer
The next parameter that was adjusted was the flow rate. There is no doubt that increasing the flow rate reduces the elution time. At some point, however, the higher flow may result in a loss of sensitivity. The vendor states that the operating flow range for the API-3000 ESI source is 40–1000 µL/min. The potential loss of sensitivity at higher flow rates was tested by repeat injections of the analytes at flow rates of 500, 750, and 1000 µL/min. The peak areas at each flow rate were then averaged and compared (Figure 2). Increasing the flow rate from 500 to 750 µL/min resulted in an average reduction in response of 21%. Carbutamide had the largest reduction, at 25%, and imipramine had the lowest reduction, at 16%. The flow rate of 1000 µL/min resulted in an average reduction in response of 34%. Carbamazepine had the largest reduction in peak area, at 43%, and verapamil had the lowest reduction, at 24%. Additional tests were performed using in-house compounds under these same conditions, and similar results were obtained. These data were subsequently used to determine the overall reduction in sensitivity and the effect on our ability to detect compounds in our absorption, distribution, metabolism, and excretion (ADME) assays. This analysis determined that moving from 500 to 1000 µL/min would have minimal impact (<5%) on the number of compounds not detected, and the benefit of reduced analytical run times far exceeded the small reduction in the percentage of compounds not detected.
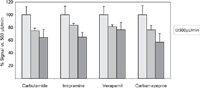
Figure 2: Comparison of signal intensity of four test solutions at multiple flow rates. Signal
A common belief is that the loss of sensitivity at higher flow rates can be overcome by splitting the flow postcolumn and doubling the injection volume. Although that may work in some applications, we found that in our case, this option yielded a signal that was 35% less than expected (data not shown). Because of those findings, flow splitting was not implemented in our methodology. Updating the method with the new flow rate of 1000 µL/min resulted in analyte retention times between 0.50 and 0.60 min (Figure 1d). The new flow rate enabled the method to end at 0.95 min (instead of the previous 1.5 min), yielding a method with an injection-to-injection time of 1.35 min (81 s).
Method Changes — Autosampler
Two modifications were considered with regard to the autosampler: the number of wash cycles and the time spent opening and closing the refrigerated sample tray door for each injection. Incorporating the previously described modifications resulted in a method of 1.35 min, which may not be enough time to complete our usual wash cycle of two needle and two valve washes. A test was performed to determine the carryover when the number of wash cycles is reduced. The wash station used the 100-µL L-MARK syringe (LEAP Technologies) that is standard with the system. Because carryover can be both compound- and system-dependent, we replaced the analyte test solution previously described with 93 in-house compounds, representing a wide range of chemicals used in our drug discovery operations. Tests were performed reducing the number of washes from four down to one, and the average carryover was compared (Table I). Wash solution A consisted of 33:33:33 (v/v/v) isopropyl alcohol–methanol–water and wash solution B was 50:50 (v/v) isopropyl alcohol–acetonitrile. The syringe filling speed for all tests was 50 µL/s and the injection speed was 25 µL/s. Each test consisted of two analyte injections followed by two blank injections (10 µL each). The four-wash condition consisted of two needle washes and two valve washes, one from each wash solution, filling the needle to its maximum capacity (100 µL). The three-wash experiment used two valve washes and only one needle wash of solution A, whereas the two-wash experiment used two valve washes only. The one-wash experiment consisted of one valve wash with solution A.
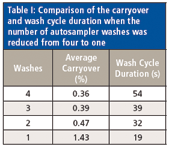
Table I: Comparison of the carryover and wash cycle duration when the number of autosampler washes was reduced from four to one
The experiments above revealed that there was minimal difference in the amount of carryover seen between the four- and three-wash conditions, 0.36 and 0.39%, respectively. However, there was a 15-s reduction in wash cycle time (54 to 39 s). The two-wash condition reduced the wash cycle time down to 32 s, but there was a slight carryover increase to an average of 0.47%. When only one wash cycle was used, the average carryover increased approximately threefold to 1.43%, but the cycle time was reduced markedly, down to only 19 s. Defining what an acceptable level of carryover would be is a topic open for debate. For in-vitro ADME studies, our laboratory prefers to see an average carryover of <1%. Considering this criteria, only the single-wash cycle would be outside our limits. Given that our injection-to-injection time was currently 57 s, we decided to use the three-wash cycle option to keep the possibility open to reducing this run time even further.
The second adjustment we made to the autosampler was the manner in which it retrieved the samples. Initially, the autosampler would open and close the refrigerated sample tray for each injection. A method that injects samples from the same plate, with samples that are thermally stable at room temperature, could save several seconds in cycle time by leaving the tray open between injections. The closing of the tray between injections is an adjustable setting on the autosampler. After the restore mode was changed from "sample" to "auto," the drawer would remain open until an injection from a different drawer was required. Although this change had no bearing on the method time, it did decrease the injection-to-injection time by ~9 s. After implementing this last modification, a method that achieved a 72-s injection-to-injection time was achieved.
Method Changes — Analytical Column
C18 was chosen as our stationary phase, with the belief that it would give us the most flexibility in retaining the diverse library of compounds we are responsible for testing. Also, because our goal was to decrease the method time, a smaller column was selected (30 mm × 2.1 mm). Our larger-capacity assays generate analytical runs as large as six 384-well plates for a single experiment, which equates to 2304 injections. Given the size of these large experiments, sample preparation is kept simple by using protein precipitation, centrifugation, and direct injection from the crash plate. The first columns tested had particle sizes smaller than 5 µm (3.5 or 2.6 µm) from various vendors. The decreased particle size resulted in higher resolution and therefore higher sensitivity based on a higher signal-to-noise ratio. However, given the relatively "dirty" samples generated from our simple sample preparation, it was discovered that the columns with smaller particle sizes did not consistently last through a 2000-injection run without clogging, causing failure of the system as a result of high back pressure. Moving to a 5-µm column largely eliminated problems with column clogging during a run and we believe it is a suitable particle size for large analytical runs using pumps not designed for ultrahigh-pressure liquid chromatography (UHPLC).
Results and Discussion
We were able to reduce our original injection-to-injection cycle time from 3.4 to 1.2 min with simple modifications of our original system components. Multiplying this reduction over a large run of 2304 injections results in a time savings of 84.5 h (130.5 vs. 46 h total run time), or 3.5 days. Over the course of the experiments summarized above, we found that even the tiniest changes resulted in considerable time savings during a run. The simple act of leaving the sample tray open between injections saved a mere 9 s. Although that doesn't sound like much at first, that small savings shaved 5.75 h from the total run time. An important fact to keep in mind is that a 46-h run time ensures that if a scientist starts the analysis on Monday, he or she could process the data on Wednesday. A run that takes longer would require the scientist to wait until the following day to process the data, thus losing a day to report results. The main reason this work was performed was to to achieve an overall reduction in the cycle time from an assay request to data released.
The 72-s method described in this article has been used for our intrinsic clearance assays. We believe these conditions provide us with a robust method that can be universally applied as a first line methodology for LC–MS-MS analysis because we are able to achieve a 94% analytical success rate over a diverse range of chemicals (with a molecular weight range from 150 to 1200 g/mol, and a cLogP range from –3 to 9). We believe the approach is easily transferable to other LC–MS-MS systems, and that similar modifications to reduce system dead volumes are possible, regardless of the HPLC instrument used. Additional modifications, such as column selection, tubing size and length, and software adjustments, could enable the method to be optimized even further.
Having the option to upgrade the mass spectrometer to a more sensitive platform would open up several new opportunities for analytical cycle time reduction. First, the overall success rate of detecting compounds would increase as a result of the higher sensitivity of the instrument. Secondly, and a more intriguing advantage would be the ability to pool samples for analysis as a result of the higher scan rates, as well as the higher sensitivity of the instrument. It is reasonable to project that the sensitivity obtained from singlet samples analyzed on an API-3000 instrument could be maintained using those same samples pooled into groups of three, four, or even six by using an API-4000 instrument for analysis. Given this scenario, one would obtain data for six analytes over 72 s, or 12 s per analyte. A 12-s per analyte process is now within the same cycle time range as the most state-of-the-art LC–MS systems currently on the market.
Conclusion
Incorporating a few simple, minor adjustments to a common LC–MS system resulted in the ability to obtain a 72-s injection-to-injection time. The system was devoid of any advanced or upgraded equipment such as column switchers, multiple HPLC systems, or aftermarket scheduling software. The system is robust, capable of long (2000+) injection sequences, and can be applied as a universal approach with a high success rate (>90%) over a diverse range of chemicals.
Acknowledgments
The authors wish to thank Sean Orlowicz of Agilent Technologies for helpful discussions regarding ways to reduce system dead volume, and Sonia de Morais, director of drug metabolism at Abbott Laboratories, for enabling our work and content review.
Lance Heinle is a Scientist II and Gary Jenkins, PhD, is a Senior Scientist II, both in Global Pharmaceutical Research and Development at Abbott Laboratories, in Abbott Park, Illinois. Please direct correspondence to: lance.heinle@abbott.com.
References
(1) S.M. Paul, D.S. Mytelka, C.T. Dunwiddie, C.C. Persinger, B.H. Munos, S.R. Lindborg, and A.L. Schacht, Nat. Rev. Drug Discov. 9 (3), 203–214 (2010).
(2) J.B. Quinn, Sloan Manage. Rev. 41, 13–28 (2000).
(3) T. Randall, "Merck, Pfizer Research Strategies Diverge on Spending," Bloomberg Bus. Week (February 3, 2011). Retrieved July 8, 2011, from www.bloomberg.com/news/2011-02-03/merck-pfizer-strategies-diverge-on-the-value-of-spending-on-drug-research.html.
(4) Rate of funding for biomedical research slowing, decreasing in recent years. ScienceDaily (January 12, 2010). Retrieved July 6, 2011, from http://www.sciencedaily.com/releases/2010/01/100112165236.htm.
(5) "Switching Between Standard and Low Delay Volume Configurations Using a Software-Controlled, High-Pressure Valve for Optimized Separation Performance," Publication Number 5989-7732EN, Agilent Technologies (Santa Clara, California, April 1, 2008).

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
The Fundamental Role of Advanced Hyphenated Techniques in Lithium-Ion Battery Research
December 4th 2024Spectroscopy spoke with Uwe Karst, a full professor at the University of Münster in the Institute of Inorganic and Analytical Chemistry, to discuss his research on hyphenated analytical techniques in battery research.
Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.