Interaction of Indigo Carmine with Nucleic Acids in the Presence of Cetyltrimethylammonium Bromide: Spectral Studies and the Confirmation of Combined Points
The interaction of indigo carmine with nucleic acids in weak acid medium was studied in the presence of cetyltrimethylammonium bromide (CTMAB) using resonance light scattering, UV–vis, and NMR.

Some organic dyes have important environmental effects; for instance, dye dilution methods for determining blood flow are used in a wide variety of circumstances (1). Most of them have low-potential toxicity and can be detected easily by ordinary spectrophotometric methods (2). However, some azo dyes can be decomposed to produce aromatic amine changing the structure of DNA, which results in human pathological changes and induces cancer (3).
Ever since its introduction in 1903 by Voelcher and Joseph, indigo carmine (IC) has been used to test renal function and it has been the subject of investigation for many years (4,5). IC usually is dispensed for intravenous injection in 0.8% of water solution in 5-cc ampules. The compound's molecular structure is shown in Figure 1.

Figure 1
Resonance light scattering (RLS) is a creative application of light signals to the measurement of RLS by simultaneously scanning the excitation and emission monochromators with a common fluorescence spectrophotometer. It has been proved that RLS has the advantages of simplicity and high sensitivity for detecting cation porphrins (6–8), triphenylmethane dyes (9), phenothiazinium dyes (10,11), xanthene dyes (12,13), metal complexes (14–16), cationic surfactant (17), zwitterionics (18), and nanoparticles (19,20). Up until now, only a few investigations have been carried out to determine nucleic acids as probe reagents for anionic dye (21–23), due to the electrostatic repulsion between nucleic acids and it.
In this study, we first find that the ternary complex of IC-cetyltrimethylammonium bromide (CTMAB)-nucleic acids can cause a stronger RLS signal. The enhanced intensity of RLS is proportional to the concentration of nucleic acids in a wide range. The sensitivity, selectivity, and linear range for the determination of nucleic acids by RLS are reported. The combined points of the anionic dye IC and nucleic acids-CTMAB have been tentatively confirmed by using 1 H NMR spectroscopy. The reaction mechanism and the influential factors on the variation of RLS have been investigated.
Experimental
Reagents
Stock solutions of nucleic acids were prepared by dissolving commercially purchased yeast DNA (Shanghai Changyan Pharmaceutical Factory, Shanghai, China), calf thymus DNA (Shanghai Changyan Pharmaceutical Factory), fish sperm DNA (fsDNA, Shanghai Institute of Biochemistry, Academy of Science, Shanghai, China), and yeast RNA (Shanghai Changyan Pharmaceutical Factory) in doubly distilled water. These stocks needed to be stored at 0–4 °C with only an occasional gentle shake if needed. The concentration of DNA was determined according to the absorbance values at 260 nm by using ε DNA = 6600 mol-1 L cm-1 (24). In this experiment, all working solutions of DNA were 25.0 mg/L.
A 1.0 X 10-3 mol/L stock solution of IC (Shanghai Chemical Reagents Co. Shanghai, China), was prepared by dissolving 0.4664 g of the crystal product in water in a 1000-mL volumetric flask. The working solution of IC was 5.0 X 10-5 mol/L.
A 5.0 X 10-4 mol/L stock solution of cation surfactant CTMAB (Merck, Germany) was prepared by dissolving 0.3645 g of the crystal product in water in a 1000-mL volumetric flask.
A 0.10 mol/L Tris-HCl buffer solution was prepared by dissolving 12.1140 g of Tris in 1000 mL volumetric flash in water and then adjusting the pH to 5.47 with 1.0 mol/L HCl.
All reagents were of analytical reagent grade without further purification and doubly distilled water was used throughout the process.
Apparatus
The RLS spectrum and the intensity of RLS were measured with an RF-5301PC (Shimadzu, Japan) fluorescence spectrometer by using a 1.00-cm quartz fluorescence cell. All absorption spectra were measured on a UV-265 spectrophotometer (Shimadzu, Japan). The surface tension (σ) was measured with a Krüss KizMK5 program surface tension instrument (Krüss GmbH, Hamburg, germany) by using the suspended plate method. 1H NMR spectra were recorded on a BRUKER 400 spectrometer (Bruker, Billerica, Massachusetts) for solution in D2O with tetramethylsilane (TMS) as internal standard. Chemical shifts are reported downfield in parts per million (ppm) and J-values are in Hz. A pHs-3C digital pH meter (Leici, Shanghai) was utilized to detect the pH values of the aqueous solutions.
Standard procedure
Into a 25-mL volumetric test tube were successively added appropriate volumes of nucleic acids solution according to the concentration needed: 1.50 mL 5.0 X 10-4 mol/L CTMAB, 1.50 mL 5.0 X 10-5 mol/L IC, and 2.00 mL HMTA-HCl buffer. The mixture was diluted to the mark with water and mixed thoroughly before RLS was measured. All measurements were made at an ambient temperature of 25 °C.
All RLS spectra were obtained by scanning simultaneously the excitation and emission monochromators (namely Δλ= 0 nm) from 250 to 700 nm. The intensity of RLS was measured at λ = 336 nm in a quartz fluorescence cell with slit width at 3.0 nm for the excitation and emission. The enhanced intensity of the CTMAB-IC system by DNA was represented as ΔIRLS = IRLS – I0 RLS. Here, IRLS and I0RLS were the intensity of the system with and without nucleic acids.
Results and Discussion
Features of the resonance light scattering spectrum
As can be seen from Figure 2 (curves 1–4), when yDNA is mixed with CTMAB, strong RLS signals could be found, because CTMAB could be aggregated on the molecular surface of nucleic acids (25) (curve 2). However, it is still much weaker than those of curve 1, which indicates that the RLS of DNA is enhanced with the help of both IC and CTMAB, and a ternary complex (26) of IC-CTMAB-DNA is produced. The CTMAB in the system acts as a bridge between IC and DNA, which results in the greatly enhanced RLS intensities. Similar RLS spectral characteristics can be surveyed when fsDNA, ctDNA, and yRNA are used.

Figure 2
The absorption spectra of the IC-CTMAB-DNA system in the 220–700 nm region are presented in Figure 3. By comparing the light-scattering and absorption spectra, it can be seen that the peaks of light scattering at 336 nm appear at the red side of the absorption bands at 292 nm. According to the theory of RLS (1,27), the peaks of light scattering at 336 nm are ascribed to the absorption bands of IC at 292 nm. Because the light-scattering spectrum depends on the nature of the system, also reflects the characteristics of the instrument (28), the RLS peak around 470 nm is considered to be possibly the emission of the xenon lamp in this region (29). The RLS intensity at 336 nm is the maximum, so 336 nm was selected in the further study.

Figure 3
Effect of buffer and surfactant
The effects of buffer and surfactant are examined. Tris is the best buffer tested, HMTA-HCl (88.7); NH4Ac-HAc (19.2); NH3-NH4Cl (11.5). It can be seen from Figure 4 that CTMAB is the best surfactant.

Figure 4
The effects of buffer and surfactant are examined (presented in Figure 4) and we know the enhanced RLS signals strongly depend upon the concentration of CTMAB. If the concentration of CTMAB is too small, the enhanced RLS intensity is not significant. However, when the concentration of CTMAB increases, the enhanced RLS signals are very strong. When the concentration of CTMAB is above 4.0 X 10-5 mol/L, perhaps CTMAB reacts with IC directly and forms larger particles of IC-CTMAB which is not favorable for further reaction with DNA, so the ΔIRLS decreases.
From the measurement of surface tension (shown in Figure 5), we can obtain the critical associate concentration (CAC) in this system, which is 3.0 X 10-5 mol/L. So 3.0 X 10-5 mol/L CTMAB is selected for further research.
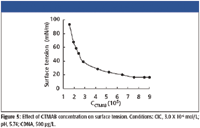
Figure 5
The effects of buffer are examined, and the dependence of light-scattering upon pH might be relevant to the form of IC and DNA. When the pH is below 4.50, both the free base form of IC and DNA are protonized, which is not favorable for positively charged CTMAB to be close to the DNA molecule. At the same time, the concentration of IC with two negative charges is low, which is not favorable for the reaction of IC and CTMAB. When the pH is above 6.50, the ΔIRLS of IC-CTMAB-DNA system begins to decline markedly, perhaps as a result of the direct reaction CTMAB with IC. So pH 5.47 is chosen for further research.
Effect of indigo carmine concentration
The effect of IC concentration on ΔIRLS of the IC-CTMAB-DNA system is investigated and shown in Figure 6. As is indicated by the figure, the IC concentration has an obvious effect on ΔIRLS of the IC-CTMAB-DNA system. It is found that when the concentration of IC is in the range 2.0 X 10-6 to 4..0 X 10-6 mol/L, the ΔIRLS of the IC-CTMAB-DNA system reaches a maximum. So we select 3.0 X 10-6 mol/L IC for further research.
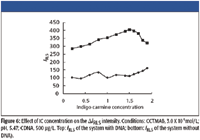
Figure 6
Effect of the ionic strength
The ionic strength of the medium has an effect on the interaction of IC, CTMAB, and DNA. As Figure 7 shows, the ΔIRLS of the IC-CTMAB-DNA system decreased slightly when NaCl was at a low concentration, while it decreased markedly with the increasing ionic strength. The reason could be that Na+ shielded the phosphorus negative charge of DNA, which weakened the combination of DNA and CTMAB, so the ΔIRLS decreased, despite the strong dependence of the enhanced light scattering of IC by DNA upon ionic strength ranges from 0 to 0.05.
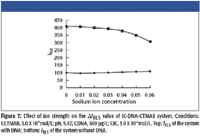
Figure 7
Reaction time and stability
The binding reaction of DNA and IC with CTMAB occurs rapidly at room temperature after less than 2 min, and the ΔIRLS remains a constant for about 4 h. It shows that the reaction does not require any crucial timing and has good stability.
Tolerance of Foreign Coexisting Substances
The effect of substances such as metal ions, amnion acids, galactose, and adenine were examined for interference. The results are summarized in Table I. It can be seen that most of the metal ions in biological systems, such as K+, Na+, and Ca2+, can be tolerated at high concentrations because they are hard ions (30) and tend to bind almost exclusively with the phosphate groups, stabilizing the Watson–Crick double helix of DNA, so their effect on the interaction of IC is related to the shielding effects of counter ions on the negative backbone of nucleic acids, which shows that this method is very useful and valuable.

Table I: Interference of foreign substances
Calibration
Under the optimum conditions, the dependence of ΔIRLS upon the concentration of DNA is determined. The analytical parameters of this method are listed in Table II, which shows that there was a good linear relationship between ΔIRLS and nucleic acids in a wide range. It is well known that no groove or helix structure exists in RNA, However, both DNA and RNA can increase the resonance light scattering intensity of IC-CTMAB in our experiment. So groove binding and intercalative binding are not the reason for the RLS enhancement.
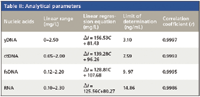
Table II: Analytical parameters
Nature of the interaction of IC, CTMAB, and nucleic acids
Generally, organic dyes interact with DNA in the following models: intercalative, groove or electrostatic binding, and the long-range assembly on the molecular surface of DNA does not involve intercalative or groove binding. It can be seen from Figure 2 that with the addition of CTMAB, DNA can enhance the ΔIRLSwithout IC, which is due to the formation of a DNA-CTMAB association, although the ΔIRLS is smaller than that in the presence of IC. IC and DNA cannot react with each other due to the electrostatic between them. By introducing a bridge, namely, cationic surfactant CTMAB into DNA-anionic dyes system, IC, CTMAB, and DNA formed a ternary complex of IC-DNA-CTMAB, which shortened the distance between IC and DNA and enhanced the assembly of IC on the surface of DNA. Several other surfactants have been employed to examine further the surfactant effects functioning in RLS enhancement. The cationic surfactants revealed RLS enhancement effects much more prominently, which in turn disclosed that electrostatic force performs an important function in the combination of DNA with IC and surfactant. The relatively weak RLS enhancement effects of non-ion and anion surfactant confirms that hydrophobic force is also a factor in the reaction of DNA with IC and surfactant (31)
It is recognized that viscosity is one of the important parameters in the reaction of DNA and small molecules (32). The relative viscosities of different mixtures were examined in the experiment and the relative viscosity of the ternary complex declining slightly indicated that there was no intercalative among CTMAB, DNA, and IC. So we can conclude that the interaction of nucleic acids, CTMAB, and IC mainly depend upon electrostatic binding. Hydrophobic force is also the factor in the reaction of nucleic acids and IC with surfactant.
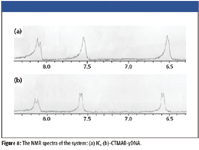
Figure 8
The NMR spectra of the system
To further understand the interaction between IC, CTMAB, and DNA, NMR spectra at optimum experimental conditions were measured, as is shown in Figure 8 and Table III. Corresponding to IC, the chemical shift signals of all hydrogen atoms moved downfield significantly when IC was added into the CTMAB and yDNA solution. The motion to downfield of the chemical shift is attributed to the reduction of the electron cloud density. It is considered that the decrease of electron cloud density of hydrogen atoms is attributed to the fact that IC combines with nucleic acid and CTMAB. The (c) hydrogen atoms are moved downfield, and this indicates the combined points are the three hydroxyl of IC through hydrogen bond and form IC-CTMAB-nucleic acids ternary complex, resulting in the RLS enhancement of this system. This is consistent with the earlier studies.

Table III: Chemical shifts (ppm) of individual protons of IC in the systems
Conclusion
In this article, a new RLS assay of nucleic acids is proposed. IC, CTMAB, and nucleic acids can form a ternary complex mainly through electrostatic binding, hydrophobic force, and hydrogen bond, which results in the enhanced DIRLS of IC-CTMAB-nucleic acids system. The combined points of the anionic dye with nucleic acids have been tentatively confirmed through the measurement of 1H NMR spectra, which establishes a foundation for further investigation on the nucleic acids at the molecular level, and have enlarged the range of probe reagents of nucleic acids at the same time. This method enlarges the range of probe reagents of nucleic acids, which will be applied widely for quantitative determination of nucleic acids. The reaction of IC and DNA directly hope to benefit the further research on the interaction of pathological changes part of DNA and the antineoplastic.
Acknowledgments
This work was supported by Xiangtan University (08XZX11).
Changqun Cai, Xiaoming Chen, and Hang Gong are with Xiangtan University, Xiangtan, Hunan, China.
References
(1) Dat1. W.L. William, U. Carlos, and V.N. Elliot, Circ. Res. 3, 570–574 (1955).
(2) K.T. Chung, S.E. Stevens, K.T. Chung, and S.E. Stevens, Environ. Toxicol. Chem. 12, 2121–2132 (1993).
(3) S.F. Chen, Y.F. Li, and C.Z. Huang, Talanta. 70, 52–57 (2006).
(4) C.Z. Huang, K.A. Li, and S.Y. Tong, Anal. Chem. 68, 2259–2264 (1996).
(5) Z.X. Guo, L. Li, H.X. Shen, and X. Cong, Anal. Chim. Acta. 379, 45–51 (1999).
(6) Z.G. Chen, W.F. Ding, Y.Z. Liang, F.L. Ren, and Y.L. Han, Microchim. Acta. 150, 35–42 (2005).
(7) C.Z. Huang, Y.F. Li, X.H.Huang, and M. Li, Anal. Lett. 34, 1117–1132 (2001).
(8) Y.T.Wang, F.L. Zhao, K.A. Li, and S.Y. Tong, Anal. Chim. Acta. 396, 75–81 (1999).
(9) C.Z. Huang, Y.F. Li, X.H. Huang, and M. Li, Analyst 125, 1267–1272 (2001).
(10) Y. Liu, C.Q. Ma, K.A. Li, and S.Y. Tong , Anal. Biochem. 268, 187–192 (1999).
(11) W.J. Zhang, H.P. Xu, C.X. Xue, X.G. Chen, and Z.D. Hu, Anal. Lett. 34, 553 (2001).
(12) Y.M. Hao and H.X. Shen, Anal. Chim. Acta. 413, 87–94 (2000).
(13) Y.M. Hao and H.X. Shen, Anal. Chim. Acta. 422, 159–166 (2000).
(14) G.W. Song, Z.X Cai, and L.Li, Microchim. Acta. 144, 23 (2004).
(15) R.T. Li, J.H. Yang, C.X. Sun, X. Wu, X.B. Gao, and Y. Liu, Microchim. Acta. 147, 105–109(2004).
(16) Z.G. Chen, W.F. Ding, F.L. Ren, J.B. Liu, Y.L. Han, and Y.Z. Liang, Anal. Chim. Acta. 550, 204–209 (2005).
(17) J. Yguerabide, G. Bee, K. Yamout, L. Korb, and J. Beck, Nat. Gen. 23, 67–68 (1999).
(18) P.Bao, A.G. Frutos, C. Greef, J. Lahiri, U. Muller, T.C. Peterson, L. Warden, and X.Y. Xie, Anal Chem. 74, 1792–1797 (2002).
(19) X.M. Chen, C.Q. Cai, J.X. Zeng, Y.Z. Liao, and H.A. Luo, Spectrochim. Acta Part A 61, 1783-1788 (2005).
(20) X.M. Chen, C.Q. Cai, H.A. Luo, and G.H. Zhang, Spectrochim. Acta Part A 61, 2215–2221 (2005).
(21) M.Wang, J.H. Yang, X. Wu, and F. Huang, Anal. Chim. Acta. 422, 151–158 (2000).
(22) B.A. Thomas and J.C. Birdsall, J. Am. Med. Ass. 69, 1747–1752 (1917).
(23) O.S. Lowsley and T.J.Kirwin, Clin. Urol. 45 (1940).
(24) Z. Chen, J. Liu, and D. Luo, Biochemistry Experiments (Chinese University of Sciences and Technology Press, Hefei, China, 1994).
(25) S.P. Liu, X.L. Hu, H.Q. Long, and L. Fan, Science in China (series B) 32, 18 (2002).
(26) R.T. Liu, J.H. Yang, X. Wu, and T. Wu, Anal. Chim. Acta. 448, 85–91 (2001).
(27) R.F. Pasternack and P.J. Collings, Science 269, 935–939 (1995).
(28) C.Z. Huang, Y.F. Li, and X.D. Liu, Anal. Chim. Acta. 375, 89–97 (1998).
(29) F.S. Collins, A. Patrinos, E. Jordan, A. Chakravi, R. Gestelan, and L.R. Walters, Science 282, 682–689 (1998).
(30) K.B. Jacobson and J. Turner, Toxicology 16, 1(1980).
(31) C.Z. Huang, X.D. Lu, and Y.F. Li, Anal. Chim. Acta. 494, 11–19 (2003).
(32) H.Li, X.Y. Yue, J.Z. Wu, and J. Liu, Acta Chim. Sinica. 61, 245 (2003).

New Study Reveals Insights into Phenol’s Behavior in Ice
April 16th 2025A new study published in Spectrochimica Acta Part A by Dominik Heger and colleagues at Masaryk University reveals that phenol's photophysical properties change significantly when frozen, potentially enabling its breakdown by sunlight in icy environments.