Simplifying Mass Spectrometry Through New Ionization Technology: Application to Drugs and Clinical Analyses
Special Issues
A newly discovered method is described for generating gas-phase ions from volatile and nonvolatile compounds. The method, matrix-assisted ionization (MAI), is both simple and sensitive, requiring only the vacuum inherent with all mass spectrometers and a suitable matrix, eliminating the need for lasers, electric fields, nebulizing gas, and even heaters to generate gas-phase ions. MAI is applicable for the direct analysis of drugs from biological fluids and tissue without prior purification. By placing matrix only on a specific surface area of interest and exposure to the vacuum of the mass spectrometer, ions are observed from compounds within the targeted surface area of tissue exposed to the matrix solution, thus allowing rapid and simple interrogation of “features of interest.” The limit of detection for drug standards is low attomoles and clean full mass range mass spectra are obtained from low femtomoles of the drug.
A newly discovered method is described for generating gas-phase ions from volatile and nonvolatile compounds. The method, matrix-assisted ionization (MAI), is both simple and sensitive, requiring only the vacuum inherent with all mass spectrometers and a suitable matrix, eliminating the need for lasers, electric fields, nebulizing gas, and even heaters to generate gas-phase ions. MAI is applicable for the direct analysis of drugs from biological fluids and tissue without prior purification. By placing matrix only on a specific surface area of interest and exposure to the vacuum of the mass spectrometer, ions are observed from compounds within the targeted surface area of tissue exposed to the matrix solution, thus allowing rapid and simple interrogation of “features of interest.” The limit of detection for drug standards is low attomoles and clean full-mass-range mass spectra are obtained from low femtomole amounts of the drug.
Mass spectrometry (MS) is a powerful analytical technology capable of accurately measuring the masses of ionized gas-phase compounds and their fragments even when present in minute quantities in complex mixtures (1). As with most technology-driven instruments, the cost and size of mass spectrometers continues to decrease while capabilities increase (2). The high performance magnetic sector mass spectrometers of the 1970s and 1980s achieved high mass-resolving power, but these instruments were often large enough to fill a big room (3). Mass spectrometers with equivalent or better mass resolution, and much higher sensitivity at high resolving power, are now the size of liquid or gas chromatographs.
The $4 billion per year MS market (4) is populated by a wide variety of instruments having mass resolution up to and exceeding m/âm 1,000,000. An attribute of many of these instruments is the ability to select ions of a desired mass for subsequent fragmentation to obtain structural information. These MS-MS or MSn instruments use older methods for fragmenting molecules such as collision-induced dissociation (CID) (5), as well as newer methods such as electron-capture dissociation (ECD) (6) and electron-transfer dissociation (ETD) (7). Ion mobility spectrometry (IMS) is now also commonly used with mass spectrometers. IMS provides an extra dimension of separation based on the size, shape, and charge of ions, along with improved dynamic range (8). IMS separations, unlike separations using gas or liquid chromatography (GC or LC), occur in the gas phase on the millisecond timescale and, because of the fast separation, can be interfaced with liquid chromatography (LC) or gas chromatography (GC) to provide an extra dimension of separation before the mass analyzer (9). However, these major advances in MS likely would not have happened without a step change in ionization technology. This is because MS measures gas-phase ions, and in the first 50 years after the discovery of MS, the primary means of achieving this measurement was thermal vaporization followed by ionization of the gas-phase molecules (10). Only volatile compounds could be analyzed by MS and therefore most biological materials were not accessible.
The introduction of electrospray ionization (ESI) (11,12) and matrix-assisted laser desorption–ionization (MALDI) (13,14) in the 1980s extended the reach of mass spectrometers to most biological materials, sparking a revolution in MS development and also in the biological sciences because of the ability to observe and identify compounds at the concentrations common in living organisms. These ionization methods have been optimized since then and modified to produce a variety of ionization techniques such as desorption electrospray ionization (DESI) (15). Alternative methods of ionizing nonvolatile compounds such as field desorption, plasma desorption, fast atom bombardment, thermospray ionization, and sonic spray ionization (10), if used at all today, are mostly practiced in research laboratories. These methods, relative to MALDI and ESI, are less commercially viable because of lower sensitivity, limited breadth of applications, and operational difficulties.
Recently, new ionization processes were discovered (16–21) that have competitive sensitivity with these older methods, even when using instrumentation optimized for ESI or MALDI, and were applicable to a wide range of compound classes including volatile and nonvolatile compounds, reduced the cost of an ion source and consumables, and were operationally robust and simple to use. Herein, we discuss one of the new ionization methods: matrix-assisted ionization (MAI). MAI requires only a special small molecule matrix compound and the vacuum inherent with all mass spectrometers. The utility of MAI is described for the analysis of drugs, even directly from biological fluids and tissue without prior sample cleanup steps. The potential for use in clinical and point-of-care diagnostics using samples obtained from a medical center laboratory is discussed.
Matrix-Assisted Ionization
Fundamental studies with inlet ionization (22), a method for producing analyte ions from solvent or solid matrix compounds using a heated inlet tube linking atmospheric pressure and the vacuum of a mass spectrometer, led to the astonishing discovery that certain small molecule matrix compounds require neither heat nor an inlet tube to produce abundant analyte ions from minute quantities of analyte (23). This ionization method was termed matrix-assisted ionization (MAI) because, like MALDI, a matrix is used and, unlike MALDI, no laser, and thus no laser desorption (LD), is required. Unlike ESI, no nebulizing or desolvation gases, heated inlet, high voltage, or clog-prone spray emitters are required (24). The only requirement is a means of exposing the matrix–analyte sample to the vacuum of the mass spectrometer, which can occur at the atmospheric pressure to vacuum inlet aperture. Either a skimmer or inlet tube can be used effectively over a temperature range from room temperature to approximately 100 °C, and in some cases from freezing conditions (25). Of course, a means of reproducibly introducing the matrix–analyte sample to the sub-atmospheric pressure is desirable. While a large variety of sample holders, including metal plates, glass capillaries, paper, and meshes, have been shown to be applicable for MAI sample introduction (26), approaches that introduce the matrix–analyte sample directly to the sub-atmospheric pressure show the greatest advance in terms of reproducibility (27).
An example of MAI is demonstrated for the analysis of a mixture of the drugs clozapine (molecular weight [MW] 326 Da), lavaquin (MW 361 Da), cocaine (MW 303 Da), fentanyl (MW 336 Da), and benzoylecgonine (MW 289 Da) using a modified Z-spray inlet of a Waters Synapt G2 mass spectrometer with 3-nitrobenzonitrile (3-NBN) as the MAI matrix. The various capabilities of this approach for direct analysis using MAI are illustrated in Figure 1. Figure 1a shows the mass spectrum of the mixture, Figure 1b the IMS separation of each component by drift time displayed in a two-dimensional (2D) plot of drift time versus mass-to-charge (m/z), Figure 1c the fragmentation of the drugs without precursor selection, and Figure 1d the combination of IMS and fragmentation without precursor selection. The vertical lines in Figure 1d are where fragment ions fall in the drift time to easily associate molecular ions and fragment ions within the drug mixture, impossible without the IMS dimension (Figure 1c). The fragment ion spectrum of cocaine extracted from the 2D plot in Figure 1d was also compared to the MS-MS spectrum of cocaine that was obtained by selecting only the m/z 304 ion for fragmentation using CID (29). In the former method, all drugs are fragmented simultaneously, while in the latter approach, each ion must be selected before fragmentation increasing the time for analysis.
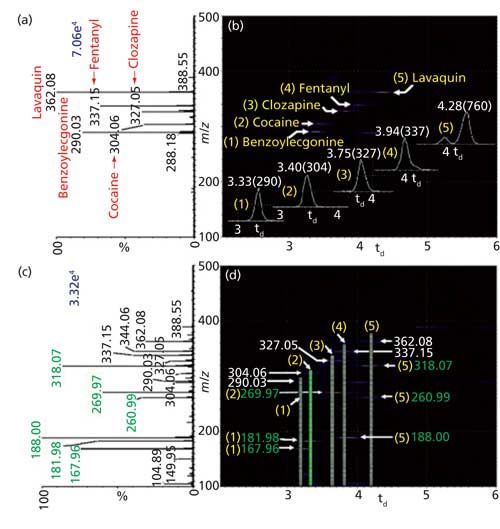
Figure 1: A drug mixture containing clozapine (MW 326 Da), lavaquin (MW 361 Da), cocaine (MW 303 Da), fentanyl (MW 336 Da), and benzoylecgonine (MW 289 Da) using 3-NBN as the MAI matrix employing a Z-spray source on a Waters Synapt G2 mass spectrometer: MAI-IMS-MS (a) total mass spectrum and (b) 2-dimensional (2D) plot of drift time versus m/z, insets show extracted drift times labeled in a nested fashion as td (m/z). MAI-IMS-MS-MS without precursor ion selection (c) total data set and (d) 2D plot of fragmentation, vertical lines highlight fragment ions for each drug. Specifically, the bright green highlight in (d) corresponding to the fragment ions is also highlighted in green color in the total dataset in (c). The extracted MS-MS spectrum is shown in the original research paper (29). Adapted with permission from reference 29.
The sensitivity of MAI is demonstrated using the MSTM platform to insert the 3-NBN matrix–analyte sample directly into the atmospheric pressure to vacuum inlet aperture of a Thermo Orbitrap Exactive mass spectrometer. The sample is a Waters MassPREP MPDS Mix 1 tryptic digest of four proteins of equal concentration. Only 50 fmol of each protein digest in the analyte mixture produces the mass spectrum shown in Figure 2. Numerous singly and multiply charged peptides are observed allowing ready Mascot software search engine (Matrix Science) identification of the yeast alcohol dehydrogenase (ADH), rabbit glycogen phosphorylase B (GPB), yeast enolase (ENO), and bovine serum albumin (BSA) proteins. The excellent sensitivity is achieved using the so called syringe method in which a few tenths of a microliter of a 1 µM aqueous solution of the digested sample is pulled into the barrel of a 1-µL syringe and expelled back into the sample container. Only solution wetting the interior of the syringe barrel is necessary for the analysis, so that only about 50 nL of the original solution is lost to the analysis. A few tenths of a microliter of 3-NBN dissolved in 3:1 acetonitrile–water is then pulled into the syringe and expelled to dry on the tip of the syringe barrel. The dry 3-NBN–analyte mixture is then inserted into the inlet aperture.
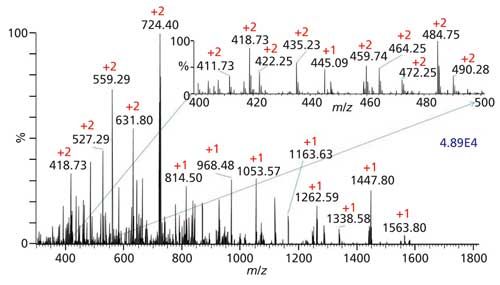
Figure 2: MAI-MS of equal concentration mixture of tryptic digest of four different proteins: yeast alcohol dehydrogenase (ADH), rabbit glycogen phosphorylase b (GPB), yeast enolase (ENO), and bovine serum albumin (BSA) proteins (Waters MassPREP MPDS Mix 1). Only 50 fmol of the analyte mixture in the MAI matrix, 3-NBN, was introduced to the inlet of the Thermo Orbitrap Exactive mass spectrometer using the syringe method on the MSTM platform.
MAI does not even require an atmospheric pressure inlet aperture, because the sample can be introduced directly into the commercial intermediate pressure MALDI source of a Waters Synapt G2 mass spectrometer. This approach was used for lipids and proteins detected directly from mouse brain tissue sections (20,28–30). However, using the MALDI sample introduction to insert sample into the intermediate pressure source is not ideal for MAI because ionization spontaneously occurs when sample is exposed to vacuum so that only a single sample can be introduced into vacuum at a time. The commercial stage for MALDI sample introduction typically requires on the order of a couple of minutes before the sample is in the ionization region and the MAI analyses can be started. While the sample is moved into position, ionization is spontaneously occurring so that many of the ions produced are lost.
To circumvent this problem, a simple probe vacuum lock was designed to allow insertion of sample into the ionization region of the intermediate-pressure MALDI housing within about 15 s. Figure 3 shows a mixture of five drugs-bromocriptine (m/z 654), buspirone (m/z 386), norfloxacin (m/z 320), oxycodone (m/z 316), and morphine (m/z 286)-introduced on the tip of a probe directly into the intermediate pressure ionization region. Each drug was present at a concentration of 0.5 µM mixed in equal volume with a solution of the matrix 3-NBN and was spotted (1 µL) on the probe tip. As shown in Figure 3, 250 fmol of each drug provides abundant analyte ions with very low background. The large differences in relative ion abundances between some of the drugs in this mixture is common in MS, as was determined using the atmospheric solids analysis probe (ASAP) method (31), which relies on vaporization followed by gas-phase ionization. In ASAP, norfloxacin is not observed and bromocriptine provides a signal-to-noise ratio (S/N) of 4 for 1 pmol loaded.
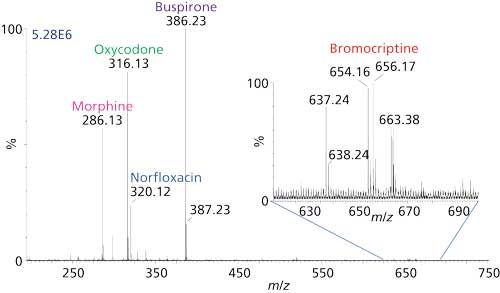
Figure 3: Intermediate pressure MAI-MS analysis of a mixture of five drugs: norfloxacin (m/z 320), bromocriptine (m/z 654), buspirone (m/z 386), oxycodone (m/z 316), and morphine (m/z 286). The 3-NBN-analyte sample was introduced using the modified probe vacuum lock on a vacuum MALDI Waters Synapt G2 mass spectrometer. Inset: zoomed view of the higher m/z range showing the MH+ ion of bromocriptine.
An additional advantage of MAI is that all matrices sublime in vacuum (32) and thus do not contribute to instrument contamination because, like solvents (18), they are pumped from the vacuum chamber. With sample inserted directly into vacuum on a probe, as discussed above, it is hypothesized that salts and other un-ionized nonvolatile materials will remain on the probe and be removed from the instrument after the analysis. Thus, MAI is potentially a very clean ionization method. Additionally, the pumping requirements of the mass spectrometer can be reduced in the absence of the continuous gas load that is necessary with atmospheric pressure ionization mass spectrometers. Pumping requirements for mass spectrometers are a major energy drain and contribute heavily to the size and weight of the instrument.
MAI will likely, over time, replace traditional ionization technologies for MS in areas where MAI offers improved performance and simplicity. Many analyses currently performed using MALDI will be accomplished with MAI in the future using common atmospheric pressure ionization mass spectrometers. Compound classes in which the MAI technology works well include drugs, metabolites, lipids, peptides, and small proteins both in positive and negative detection modes (27,29,33–37). A great benefit, especially relative to ESI, is the ability to ionize samples directly from dirty conditions without sample cleanup protocols (38). An example is shown in Figure 4 of a 24-h time-course study of Allegra Allergy (fexofenadine, MW 501 Da) analyzed directly from urine with no cleanup and only diluted 4:1 with water before analysis. Using the syringe method previously discussed, the time to acquire each sample is about 30 s. The ion abundances for the active ingredient of fexofenadine can be seen to increase in the urine and reach a steady state in about 1 h after consumption, then decreased substantially after 24 h. On the other hand, azithromycin, an antibiotic designed to remain in the body for at least 10 days after the last dose, is readily seen using the same approach 12 days after the final dose (Figure 5). These analyses would be substantially more difficult using ESI because of the salty conditions. In fact, 1 µL of a mixture of five drugs in urine could be analyzed directly using MAI or MALDI, but ESI required that the urine be diluted 20:1 with water to obtain results by summing the spectra acquired electrospraying the entire sample. For this sample, MAI provided superior results (27). Proteins, including toxins, bacteriorhodopsin, and ebola protein are converted to gas-phase ions using MAI matrices, even when they are dissolved in buffers or ionized directly from whole blood; those conditions are not amenable to ESI (20,39,40). Similarly, a proteomics approach of a digested area of tissue directly characterized by MAI-IMS-MS-MS sequencing using CID identified a peptide of tubulin beta chain from the targeted area with a Mascot score of 101 (40). At this early stage of MAI development, many classes of compounds have not yet been attempted and the upper mass range has not been explored. The largest protein analyzed at this time is BSA at 66 kDa (20).
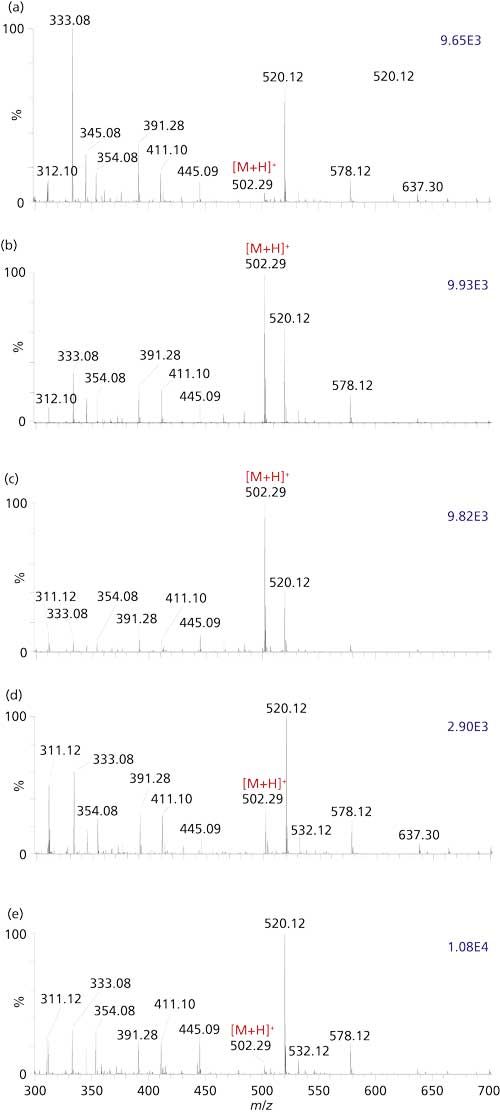
Figure 4: MAI-MS analysis of urine obtained at different time intervals after consumption of a 24-h Allegra Allergy pill. (a) 20 min, (b) 50 min, (c) 350 min, (d) 21 h and 20 min and (e) 24 h with detection of the active ingredient fexofenadine (MW 501 Da) from urine (diluted 1:4 urine-water) with no prior cleanup. The MSTM syringe method was used on a Thermo Orbitrap Exactive mass spectrometer.
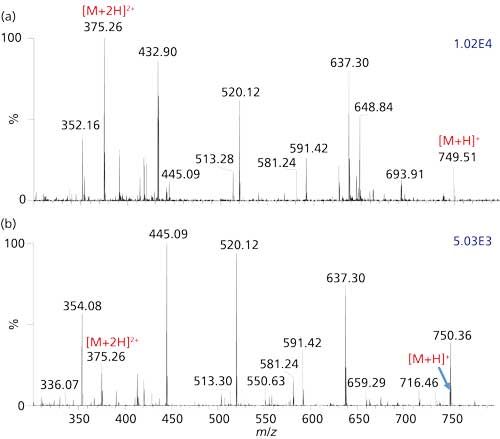
Figure 5: Detection of the antibiotic azithromycin from a urine sample excreted (a) after four days (b) after 12 days after final dose, using the MSTM syringe MAI-MS method on a Thermo Orbitrap Exactive mass spectrometer.
MAI will not supplant LC–ESI-MS, but as MS capabilities advance, direct analyses become more attractive because of speed and simplicity relative to the additional separation step (29,38,41). A spate of ionization approaches that are based on direct ionization have been introduced over the past decade, including ASAP, nanospray desorption ionization (nano-DESI), direct analysis in real time (DART), laser ablation electrospray ionization (LAESI), and liquid extraction surface analysis (LESA) (41). These methods are typically used with minimal sample workup and without a prior chromatographic separation step, although they can be interfaced with IMS-MS. None of these methods, however, are operationally as simple as MAI nor do they match the sensitivity, mass range, or the minute background.
Proof-of-principle for analyses performed directly from 96-well plates (28), thin-layer chromatography (TLC) plates (28), as well as operation in a rapid (24,26,39) and automated (Figure 6) fashion has been demonstrated. All that was required to convert an Advion Nanomate to automated MAI was 3-NBN matrix and a short extension connecting the mass spectrometer inlet with the matrix–analyte delivered to the extension by pipet tip. From the total ion current plot, it is evident that much faster analyses are possible.
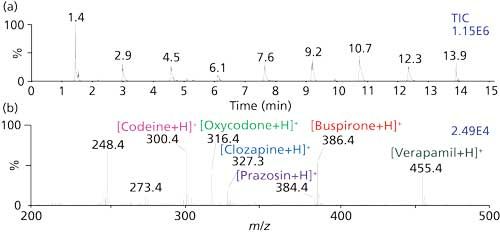
Figure 6: Automated MAI-MS analysis of six drug standards plus a urine sample spiked with the six drugs (peak at 1.4 min) using a TriVersa Nanomate ESI system (Advion) with a Thermo LTQ XL mass spectrometer. (a) Total ion chronogram for the analysis and (b) mass spectrum of the undiluted urine spiked with the six drugs.
Potential for Clinical Applications
Identifying clinically relevant biomarkers of human disease is often like searching for the proverbial needle in a haystack. MS approaches are well suited for such a search because MS interrogates compounds within complex biological samples at the molecular level with superb sensitivity, mass resolution, and mass accuracy (42,43). However, improved sensitivity and dynamic range, as well as reproducibility from sample-to-sample are required for many clinically relevant applications. The problem is exacerbated by the extreme complexity that comes with clinical samples and the need for accuracy, speed, safety protocols, and cost effectiveness. Although tandem MS targeted screening, especially when interfaced with a prior chromatographic separation method, has been successful (44), a less costly approach in time, expertise, consumables, and equipment would be a driver for MS in clinical research. As a result, we have begun to look at the potential of MAI to work with clinically relevant problems together with the Detroit Medical Center (DMC) in Detroit, Michigan.
Three factors that are important in clinical settings are quantification, reproducibility, and the absence of instrument contamination. Reproducibility and quantification were determined for a set of six small-molecule drugs using the MAI method with sample introduction to a Thermo LTQ Velos mass spectrometer using the syringe sample introduction method on a MSTM platform as well as hand injection using a pipette tip on a Waters Synapt G2 mass spectrometer (27). Reproducibility and quantification using MAI was compared to ESI on the same instruments and also vacuum MAI relative to MALDI on the Synapt G2 system. The reproducibility and quantification using MAI was competitive with ESI and somewhat better than MALDI. Using the same mass spectrometers, the potential issue of contamination was also addressed by inserting 250 MAI samples each, 500 total, of the drug buspirone and the protein ubiquitin into the LTQ Velos and Synapt G2 mass spectrometer inlets while monitoring the ion abundance of the analyte ions (Figure 7). No loss of sensitivity was observed over the course of the analysis, thus demonstrating the robustness of the MAI method.
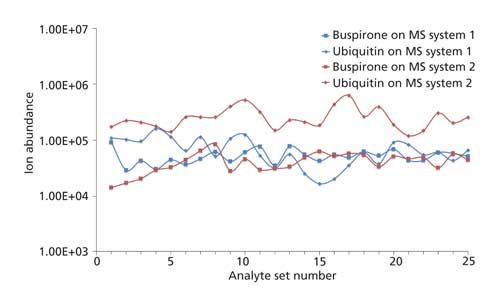
Figure 7: Plot of analyte ion abundance averaged for 10 consecutive acquisitions for the drug buspirone (1 ppm in water) and the protein ubiquitin (1 pmol/µL in 1:1 methanol–water, 1% acetic acid) over 250 injections each, alternating solutions every 10 acquisitions for a total of 500 injections on each instrument. The graph shows 25 sets of 10 acquisitions averaged to produce a single data point for each analyte. 3-NBN was used as the matrix with the inlet temperature set to 50 °C on system 1 (Synapt G2) and 150 °C on system 2 (LTQ Velos). 1 μL of dried matrix-analyte solution was introduced to the inlet aperture via pipette tip on system 1 and using the MSTM prototype platform on system 2.
Urine samples from the DMC hospital lab that had already been tested for drugs of abuse were analyzed using MAI with the 3-NBN matrix (27). The sample showed a prominent signal at m/z 300, which was suspected to be either codeine or hydrocodone. The protonated molecular ion at m/z 300 was fragmented by CID and compared to the fragmentation spectra of hydrocodone and codeine to confirm the presence of hydrocodone. The concentration of hydrocodone in the urine sample was subsequently quantified using standard addition with hydrocodone-D6 (1 ppm) as an internal standard. A linear regression calibration curve was created from the mean values of five measurements at five concentrations. Using this simple MAI-MS approach, the hydrocodone concentration in the drug-addicted-infant urine was determined to be 3.32 ppm. The relative standard deviation was found to be in the range of 2.2–5.4%, which is notably better than the typically required 20% for clinical applications (27). These results are in excellent agreement with traditional methods (LC–MS) performed in the DMC laboratories. MAI-MS was thus effective in rapidly identifying hydrocodone in the urine of an infant patient whose mother had abused opioids during pregnancy (27). This particular sample, however, contained a relatively high concentration (3.32 ppm) of hydrocodone, over two orders of magnitude above the limit of quantification for MAI using pure standards. Therefore, a similar analysis will be expanded to evaluate the ability of MAI-MS to quantify opioids at parts-per-billion levels in clinically relevant samples.
These initial MAI studies using patient urine samples reveal the diagnostic utility of MAI-MS for the detection of drugs of abuse and prescription drugs and their metabolites (29,39). An example is the ready detection of the diagnostic nicotine metabolites cotinine and nornicotine in the urine of a smoker without sample cleanup (29). Another study demonstrated the ability to quantify clozapine directly from mouse brain tissue by simply spotting the matrix on the tissue and exposing it to the vacuum of the mass spectrometer (38). The inclusion of IMS-MS with MAI cleanly separated a mixture of six drugs by IMS, and MS-MS provided almost instantaneous structural information to aid characterization without precursor selection by including the IMS drift time dimension (26,29). Although MS methods are currently available to detect drugs of abuse using LC–MS-MS (42,44,45), ambient ionization (46), or MALDI (47), the process is greatly simplified using MAI (26,27,29,38–40), which eliminates the need for expensive specialized instrumentation as in MALDI or nebulizing and desolvation gases and high voltage supplies in ESI.
Of course, there are advantages in interfacing an LC or GC to a mass spectrometer and in using mass spectrometers with high resolving power, MS-MS, and IMS capabilities for clinical analyses, but the necessary operator expertise and the cost of these instruments is prohibitive in many settings. Multiple MS instrument manufacturers have developed small, inexpensive mass spectrometers that are commercially available. Smaller, lighter, and less energy intensive mass spectrometers are currently being developed for near real-time analysis in non-laboratory settings (48–50). Because ionization using MAI occurs independent of high voltage, laser ablation, heat, or desolvation gases, the technology is portable (51) and therefore potentially amenable for use in point-of-care settings, field hospitals, or airport security screens. To access the potential of portable mass spectrometers in a clinical laboratory and establish the relative effectiveness of a low-resolution mass spectrometer as a diagnostic tool using MAI, a Waters QDa mass spectrometer was transported on a cart to the DMC. That system starts up quickly using power from standard 120-V wall sockets. The tolerance of salts is demonstrated in complex biological samples using this portable mass spectrometer with MAI as shown for hydrocodone-spiked undiluted urine (51) (Figure 8).
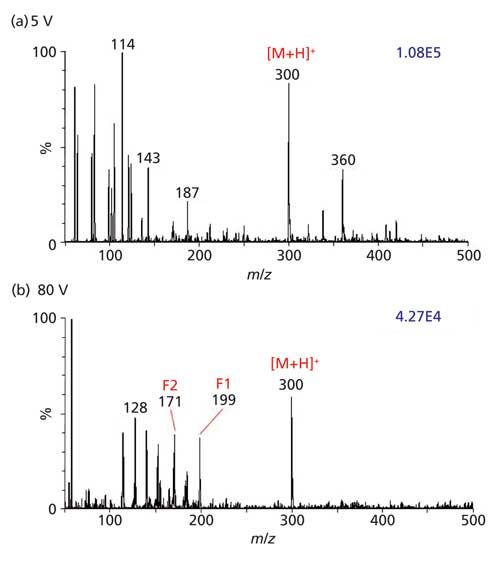
Figure 8: Characterization of 6.0 µg/mL hydrocodone drug (MW 299 Da) in urine by MAI-MS on a portable Waters QDa mass spectrometer in the DMC hospital laboratory. For MS analysis by the hospital personnel, undiluted urine was mixed 1:1 by volume with 3-NBN matrix solution. A 1-µL volume of matrix–analyte solution was crystallized on the end of a pipette tip and introduced to the inlet aperture of the mass spectrometer held at 50 °C source block temperature. Mass spectra of hydrocodone [M+H]+ ion were acquired with cone voltages of (a) 5 V and (b) 80 V. Characteristic in-source fragment ions are labeled as F1 and F2 according to literature. Adapted with permission from reference 51.
Future Outlook
MAI-MS is clearly applicable for research and industrial analysis problems. The approach offers a new ionization approach for problems not readily addressed by current technologies, as well as simplicity, sensitivity, and cost effectiveness. Future applications will capitalize on the sensitivity of MAI ionization to generate multivariate statistical models using MAI-MS and MS-MS datasets obtained from clinical specimens to aid in accurate diagnosis of disease. A fully automated MAI-MS interface for use in laboratory settings will serve to improve clinical decision making and overall response times. The ability of MAI to convert both volatile and nonvolatile compounds efficiently into gas-phase ions requiring little operator expertise and without the need for lasers, desolvation-nebulizing gases, high voltages, or a sophisticated ion source bodes well for this method in extending MS to point-of-care diagnostics. Accordingly, this simple ionization process has the potential to be a driver in making bedside MS data acquisition a real possibility.
Acknowledgment
The authors gratefully acknowledge the support from NSF CHE-1411376 to ST, NSF STTR Phase II Award 1556043 to MSTM, LLC, grants to support the Core (and to PMS) P30 ES020957, P30 CA022453, and S10 OD010700.
References
- A. El-Aneed, A. Cohen, and J. Banoub, Appl. Spectros. Rev.44, 210–230 (2009).
- L. Van Oudenhove, and B. Devreese, Appl. Microbiol. Biotechnol.97, 4749–4762 (2013).
- S.M. Press, Current Trends in Mass Spectrometry11(3), 8–11 (2013).
- D.M. Desiderio and I. Katakuse, Anal. Biochem.129, 425–429 (1983).
- R.A. Zubarev, N.L. Kelleher, and M. McCoustrat, J. Am. Chem. Soc.120, 3265–3266 (1998).
- L.M. Mikesh, B. Ueberheide, A. Chi, J.J. Coon, J.E. Syka, J. Shabanowitz, and D.F. Hunt, Biochim. Biophys. Acta1764, 1811–1822 (2006).
- X. Liu, S.J. Valentine, M.D. Plasencia, S. Trimpin, S. Naylor, and D.E. Clemmer, J. Am. Soc. Mass Spectrom.18, 1249–1264 (2007).
- S.J. Valentine, M. Kulchania, C.A. Srebalus Barnes, and D.E. Clemmer, Int. J. Mass Spectrom.212, 97–109 (2001).
- C.N. McEwen and B.S. Larsen, Int. J. Mass Spectrom. 377, 515–531 (2015).
- M. Yamashita and J.B. Fenn, J. Phys. Chem. 88, 4451–4459 (1984).
- J.B. Fenn, M. Mann, C.K. Meng, S.F. Wong, and C.M. Whitehouse, Science246, 64–67 (1989).
- M. Karas, D. Bachmann, U. Bahr, and F. Hillenkamp, Int. J. Mass Spectrom.78, 53–68 (1987).
- M. Karas, U. Bahr, and F. Hillenkamp, Angew. Chem., Int. Ed. 760–761 (1989).

Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.