The Use of MALDI Imaging Mass Spectrometry for Interdisciplinary Research: From Metabolomics to Pesticides
Special Issues
Matrix-assisted laser desorption–ionization (MALDI) imaging mass spectrometry allows direct, in situ, label-free measurement of proteins, peptides, lipids, small-molecule drugs and their metabolites, and other chemicals in tissues. In a range of applications, the unique information generated by MALDI imaging can make a significant contribution to understanding factors such as molecular and metabolic mechanisms and the transport and localization of compounds or metabolites with human, animal, or plant species.
Matrix-assisted laser desorption–ionization (MALDI) imaging mass spectrometry allows direct, in situ, label-free measurement of proteins, peptides, lipids, small-molecule drugs and their metabolites, and other chemicals in tissues. Applications range from fundamental biological research, through environmental and toxicological science, to pharmaceutical R&D. In each case, the unique information generated by MALDI imaging has made a significant contribution to understanding important factors such as molecular and metabolic mechanisms and the transport and localization of compounds or metabolites with human, animal, or plant species. MALDI imaging can also elucidate the impact of pesticides like chlordecone on human health over the long term, and provide supporting evidence for environmental legislation as well as political and economic decisions about environmental remediation and policy.
Over the past decade, the demand for highly sensitive and ultrafast mass spectrometry (MS) techniques has soared, driving the innovation of a diverse array of mass analyzers for use in a broad range of applications, such as drug discovery, 'omics studies, drug metabolism studies, and environmental sciences. Mass spectrometers are now commonplace outside of traditional analytical laboratories and the emergence of different ionization sources-such as direct matrix-assisted laser desorption–ionization (MALDI)-has propelled many advances, including in MS imaging.
MALDI imaging, a technique that was first highlighted in the late 1990s by Caprioli (1) as a sensitive tool for the study of biochemical processes in mammalian tissue sections, has made significant contributions to the field of molecular biology, particularly in the study of the transport and localization of compounds and metabolites in biological systems, including humans, animals, and plants.
Integrating Biology and Chemistry
MALDI imaging allows direct, in situ, label-free measurement of proteins, peptides, lipids, small-molecule drugs and their metabolites, and other chemicals in tissues. Early experiments by Caprioli demonstrated MALDI ion image analysis of regions of rat pancreas and pituitary. Localized peptides were mapped, and the authors noted that in a single spectrum from a rat pituitary print, more than 50 ions corresponding to the peptides present in the tissue were observed, including precursors, isoforms, and metabolic fragments (1).
More recently, a comprehensive review of the potential of MALDI imaging for the toxicological evaluation of environmental pollutants was published (2). The authors concluded that MALDI imaging has great potential for studies involving the distribution and metabolism of chemicals released into the environment, and to better elucidate their mechanisms of action.
A range of mass spectrometers can be integrated into a MALDI imaging system, depending on application needs. MALDI time-of-flight (TOF) MS, for example, offers high throughput (with lower ability to distinguish molecules of similar molecular weight), while magnetic resonance mass spectrometry (MALDI-MRMS) provides high measurement accuracy and mass resolving power.
MALDI Imaging in Pharmaceutical R&D and Cancer Research
Throughout the 2000s, MALDI imaging developed along two parallel tracks: Interest in its use as a research tool continued to grow among biologists, and at the same time, the first publications of its application in pharmaceutical research and development (R&D) were appearing.
Traditional approaches in pharmaceutical R&D for the risk assessment of safe and efficacious drugs have been based on measuring the parent drug in plasma, both in animal models and humans. It is recognized, however, that most drug targets are not located in plasma, and determining the relevant tissue distribution of not only the parent drug, but also its metabolites, would provide much greater understanding of pharmacology and toxicology.
The development of current best practice methods for quantitative whole-body autoradiography (QWBA) and liquid chromatography coupled with mass spectrometry (LC–MS) clearly move the emphasis to analyzing tissues rather than plasma, but still fall short of providing a complete picture. Both techniques have challenges. QWBA is a robust method, and the data generated are accepted by regulatory bodies around the world. However, QWBA requires the use of radioactively labeled probes and presents a composite of the total radioactivity present in the body, including any combination of parent drug, metabolites, impurities, and degradation products.
LC–MS analysis is performed on extracts from tissue homogenates. The technique cannot provide any spatial information and, importantly, can be misleading. For example, if an analyte in the tissue is highly localized, the extraction and homogenization process will act as a dilution, masking this distribution and showing a relatively low concentration, sometimes even below the limit of detection.
Against this background, MALDI imaging offers potential advantages for drug development. For example, MALDI imaging is generating important new information about the localization of drug compounds and important metabolites in tissue samples. Figure 1 shows an extract from an early work by Castellino and colleagues in which a small area of a histology section is magnified, showing regions of inflammation (3). An ion map (with 50-µm spatial resolution), which corresponds to the same region, indicates that the localization of lapatinib metabolite, M10, is only in the regions associated with the inflammation.
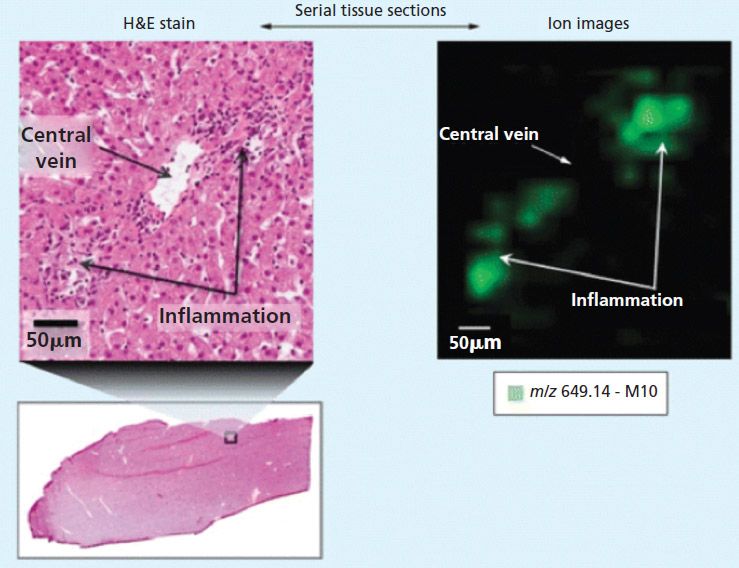
Figure 1: Correlation of histology and MALDI imaging in dog liver sections. Adapted with permission from reference 3.
In addition, MALDI imaging is contributing to the development of new biomarkers of disease, particularly in the field of cancer research. Here, the heterogeneous nature of cancer samples means that because the spatial distribution and the histology of the samples is preserved, MALDI imaging is well suited to this work. Importantly, MALDI imaging has been successfully applied to the detection of previously unknown proteins in cancer, as well as for recent advances in the biomarker discovery of N- and O-glycosylation of proteins. In such studies, MALDI imaging has been used to develop tumor-specific glycan biomarkers; this work shows promise for the characterization of N-linked glycans in cancer tissues (4).
Instrument Advances Drive New Frontiers in Biology
Importantly, although the most well-known association of MALDI imaging is in pharmaceutical applications, that industry represents just one sector that adopted the technique early. At around the same time that initial work in the pharmaceutical industry took place, several institutes in France were using MALDI techniques to gain insights into the effects of environmental toxins on reproduction, chemical interactions and structures in the food supply, and evaluating plants engineered for bioremediation of contaminated lands.
One group, led by Charles Pineau, has been using MALDI imaging and applying it to research developmental and reproductive biology and environmental and chemical toxicity. Pineau is a research director at Inserm (the French National Institute of Health and Medical Research) and a team leader at IRSET (the Research Institute for Environmental and Occupational Health). As part of his role at IRSET, Pineau is the director of Protim, a core facility involved in optimization and validation of new technologies in two fields of proteomics: extensive proteome characterization of complex samples, and MALDI imaging for toxicology and clinics.
Collaborations Powering Imaging Excellence
Protim is involved in two collaborative European Union (EU)–funded projects: "3D MASSOMICS" and "METASPACE."
The recently completed research project "3D-MASSOMICS" was funded as part of the EU FP7 HEALTH program, with the aim to enable three-dimensional (3D) label-free proteomics and metabolomics imaging using MALDI imaging (5). Traditional 3D imaging techniques such as computed tomography (CT), magnetic resonsance imaging (MRI), and positron emission tomography (PET) cannot be used for proteomics or metabolomics discovery studies, nor for biomarker discovery and disease pathway analysis. To address this challenge, the consortium united nine partners, including Protim, from six countries both from industry and research, to develop statistical methods for the reproducible collection of 3D MALDI imaging data, and for unsupervised and supervised statistical analysis and interpretation of this data.
A review by the coordinators of the consortium, Palmer and Alexandrov, aimed to assess and increase the knowledge of 3D imaging MS in the analytical community, to accelerate the innovation of this molecular imaging tool (6). The review identified that the reduction of acquisition time is important in 3D imaging MS development. When using modern MS with fast analyzers (such as TOF or quadrupole time-of-flight [QTOF] technologies) and high-frequency lasers of 2–5 kHz, the pixel-by-pixel method of data acquisition requiring high-
acceleration high-precision positioning stage becomes a bottleneck. Advanced high-performance imaging MS was reported that uses 5–40 kHz lasers, providing an acquisition speed of 20 pixels per second.
METASPACE, funded as part of the EU's Horizon 2020 research and innovation program, is an ongoing project for Protim (7). It is a specialist platform that hosts an engine for metabolite annotation of MALDI imaging data as well as a spatial metabolite knowledge base of the metabolites from hundreds of public datasets provided by the community. The METASPACE platform is developed by software engineers, data scientists, and MS experts from the Alexandrov team at the European Molecular Biology Laboratory (EMBL). As part of this project, imaging MS was used for spatial metabolomics analysis with the aim to create a bioinformatics tool for automated metabolite annotation, in high mass resolution imaging MS (8). The development of an algorithm to efficiently mine 10–100 gigabyte datasets was established, and the results of the annotation are shown in Figure 2.
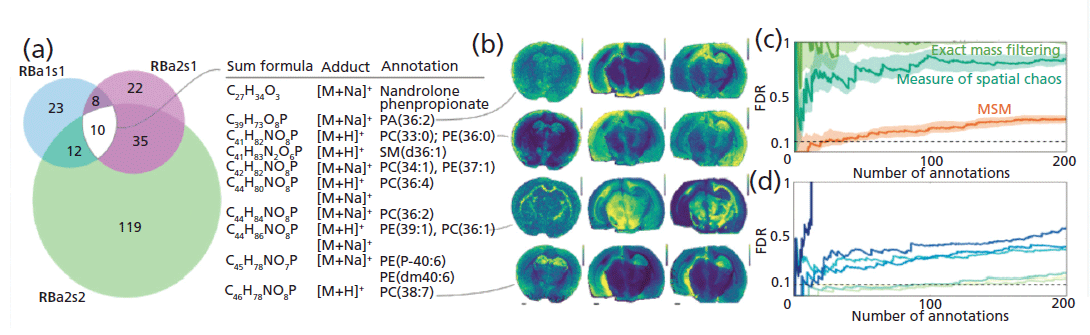
Figure 2: The FDR-controlled molecular annotation for three MALDI FT-ICR imaging MS datasets from rat brain coronal sections (HMDB, FDR desired=0.1): (a) Venn diagrams showing numbers of molecular annotations and a list of 10 annotations from all three datasets. (b) Exemplary ion images of four annotations (see Table SD1.2 in reference 8 for detailed information about all 10 annotations), as well as FDR curves illustrating: (c) superiority of MSM as compared to individually considered exact mass filtering and measure of spatial chaos, and (d) decrease of reliability of molecular annotation while simulating deteriorated mass accuracy and resolution by taking signals with a larger m/z tolerance. Adapted with permission from reference 8.
Analysis of Human Exposure to Chlordecone
In addition to the 3D-MASSOMICS and METASPACE projects, Pineau's team at Protim has a number of active research projects investigating how exposure to chemicals in the environment impacts human health. Traditionally, limits and guidelines from the U.S. Environmental Protection Agency (EPA) and other official bodies regulate acceptable environmental levels of toxic chemicals. Depending on the toxin type and matrix (air, water, or soil), compliance with these limits is typically measured using dedicated environmental chemistry methods (ECMs) such as gas chromatography (GC) and LC, often in tandem with electron-capture detection (ECD) methods (a full list of techniques and ECM methods can be found in reference 9). However, those measurements do not shed light on the mechanisms and potential impact on human health that these substances in the environment might have.
Several IRSET teams in Rennes, France, are working on analysis of the pesticide chlordecone, which was used in Martinique and Guadeloupe to combat banana weevil until 1993. Chlordecone is a known endocrine disrupter and carcinogen, and its use has been banned worldwide since 1990. Before the ban, a significant proportion of the population in the French West Indies was exposed to the chemical and the soil and coastal waters were polluted.
Initial work by epidemiologist Luc Multigner (a team leader at the IRSET institute), published in 2010, confirmed that chlordecone was responsible for a significant increase in the risk of prostate cancer, the cause of half the cancers detected on the two islands (10).
In parallel, the group at Protim was progressing new laser technology to improve the resolution of MALDI imaging. A breakthrough publication in 2011 showed for the first time that it was possible to acquire MALDI images of proteins in the 10 kDa range at 20-µm lateral resolution on a commercial instrument (11). Rat testis was used as a relevant model to demonstrate that this improved resolution corresponded to complex anatomical features at the cellular level. The correlation of molecular signals was successfully established with the development of germ cells within the seminiferous epithelium.
This level of sensitivity was deemed essential to the next stage of the chlordecone research and the work to investigate the localization of chlordecone in the body and quantify it directly on the tissue sample. The development of MALDI imaging for toxicological evaluation of small molecules requires solid quantification methods. However, such quantification is complicated with MALDI imaging for several reasons. First, endogenous species present in a tissue section can affect the intensity of the ion of interest and can lead to varying ionization efficiency across the section. Second, the heterogeneity of matrix crystallization can induce signal variations. Finally, depending on the physicochemical properties of the studied molecule and the composition of the matrix solution, the extraction of the compound of interest occurring during matrix application can vary in efficiency.
Pineau's team was able to develop a robust method for in situ quantification directly on the tissue sections, published in 2014 (12). Figure 3 shows the in situ absolute quantification method (ISAQ) of chlordecone that was demonstrated in the pathological liver of a chlordecone- and CCl4-treated mouse.
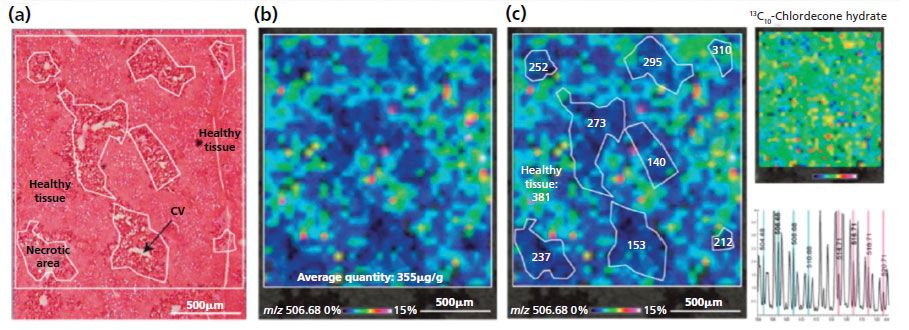
Figure 3: In situ absolute quantification of chlordecone in the pathological liver of a chlordecone and CCl4 treated mouse. (a) Microscopic image of a serial section stained with H&E. (b) MALDI image corresponding to chlordecone hydrate: an average absolute quantity of chlordecone of 355 µg/g was measured in the analyzed area. (c) Local absolute quantities measured in each necrotic areas are lower than those measured in the healthy tissue (381 µg/g). The uniform distribution of the internal standard (m/z 516.71) on the tissue section confirms that the differential distribution of chlordecone is not a result of ion suppression effects.
The group successfully developed a method for the quantification of small molecules by MALDI imaging, based on the combination of labeled normalization with the calculation of a correlation curve between MALDI imaging and GC as a conventional quantification technique. This method was successfully applied to the quantification of chlordecone in the mouse liver, and is the first example of the application of MALDI imaging to the quantification of a pesticide in tissues.
Although this study represents an important step forward in the capabilities of MALDI imaging, chlordecone has a half-life of 30 years, which means that the environment in Guadaloupe is predicted to be polluted for more than 500 years to come. Scientists continue to monitor various population groups on the islands to assess the ongoing impact on male fertility and child development and behavior. For example, studies of the effects of chlordecone exposure on pregnancy duration and the risk of preterm birth found that maternal exposure to chlordecone was strongly associated with a shorter pregnancy duration and an increased risk of preterm birth, regardless of the method of onset of labor (spontaneous or induced). The hormonal, estrogenic, and progestogenic properties of chlordecone are thought to be the cause of this association (13).
Using 3D Imaging to Advance Knowledge of Spermatogenesis
In addition to the work on the impact of chlordecone and other toxic compounds (bisphenol A and glyphosate, for example), Pineau and the Protim team are contributing to wider applications of MALDI imaging in human reproductive biology. With more than 30 years of experience, Pineau is a renowned specialist in spermatogenesis. He has made significant contributions to the development of proteomics and integrative proteomics technologies and their application to answer biological and clinically relevant problems in the field of testicular pathophysiology and reproductive toxicology. Now, MALDI imaging is playing a key role in his further investigations of the complex process of sperm maturation.
The epididymis is a small organ located next to the testicle. Immature spermatozoa are formed in the testis and are morphologically complete, but lack functionality. Gametes transit through the epididymis, acquiring motility and the ability to fertilize during this maturation process. Nuclear gene transcription is switched off at the sperm maturation level, and the process is driven by an array of sequential protein modifications that are inextricably linked to the complex epididymal luminal microenvironment. When a spermatozoon passes from caput to cauda, numerous biochemical events occur in its subdomains, including post-translational modifications, proteolytic processing, protein redistribution and disappearance, and integration of new components, thus leading to a dynamic remodeling of the sperm surface.
The unique ability of MALDI imaging to visualize molecules in situ has significantly advanced the study of this complex biology. Figure 4 shows a two-dimensional (2D) image of a rat epididymis head with highlighted peptides possibly involved in the maturation of sperm during its transit through the organ (14). Figure 5 illustrates the latest work in Pineau's lab: the creation of 3D images (15). The process of capturing a 3D MALDI imaging model of an organ begins with making many hundreds (or thousands) of sections through the organ. Each section is measured and the data are combined in software to create the final picture. The resulting data sets are huge-in the terabyte range-but once completed, a dataset can be mined for information for many years.
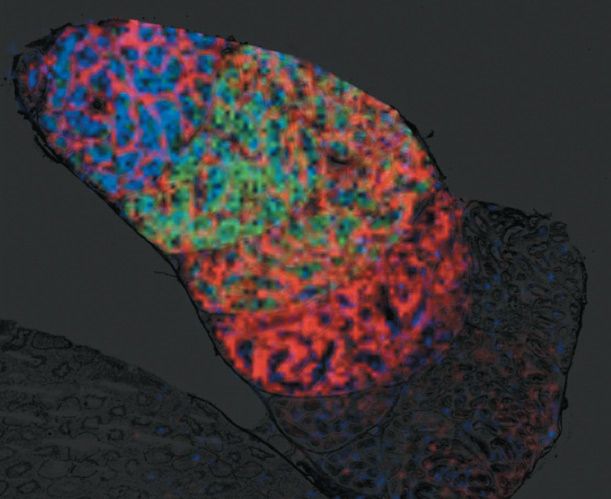
Figure 4: Molecular image of sperm transit in the rat epididymis by MALDI imaging MS. Peptides specific to maturing spermatozoa during epididymal transit can be visualized simultaneously. Signal overlay of three specific peptides corresponding to m/z 5470, 6177, and 18,746 is visualized in different colors. An 80-µm lateral image resolution is mandatory to resolve the organ structures.
Summary
MALDI imaging has emerged as a technique that offers deep understanding of the localized distribution of peptides, proteins, and small molecules in tissues. Applications range from fundamental biological research, through environmental and toxicological science, to pharmaceutical R&D. In each case, the unique information generated by MALDI imaging has made a significant contribution to understanding important factors such as molecular and metabolic mechanisms and the transport and localization of compounds or metabolites with human, animal, or plant species. As highlighted in this article, MALDI imaging can also show how pesticides like chlordecone impact human health over the long term, and provides supporting evidence for environmental legislation as well as political and economic decisions about remediation and environmental policy. Arguably still in its infancy, continuous improvements in MALDI imaging systems have made the technique more accessible, more reliable, and easier to use, and data analysis has become both more sophisticated and more automated. MALDI imaging looks set to play an increasingly important role in the coming years.
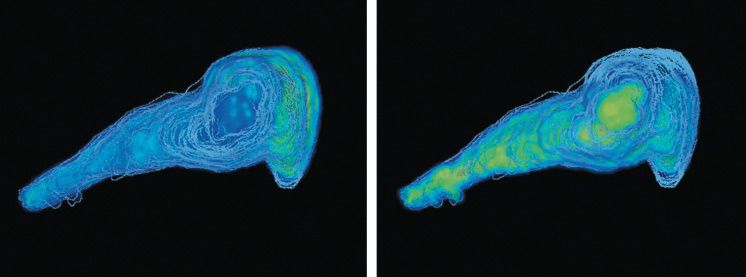
Figure 5: Three-dimensional mass spectrometry imaging view of a rat epididymis head. PLSA computation in SCiLS software with restricted mass range (m/z 250-1650). Left: m/z corresponding to a metabolite localized in the vas efferens area. Right: m/z corresponding to a metabolite expressed along the tubule.
Acknowledgments
This work was supported in part by Charles Pineau, PhD, the research director of Inserm and the director of the Protim Core Facility; Richard M. Caprioli, a professor in the Departments of Biochemistry, Chemistry, Pharmacology, and Medicine at Vanderbilt University School of Medicine, in Nashville, Tennessee; Hélène Rogniaux, PhD, a research engineer at the Biopolymères, Interactions, Assemblages (BIA) unit of the French National Institute for Agricultural Research (INRA); and Dimitri Heintz, PhD, the head of the Platform Imaging Mass Spectrometry PIMS Metabolomics laboratory (IBMP) in Strasbourg, France.
References
(1) R.M. Caprioli, T.B. Farmer, and J. Gile, Anal. Chem. 69(23), 4751–4760 (1997).
(2) M. Lagarrigue, R.M. Caprioli, and C. Pineau, J. Proteomics 144, 133–139 (2016).
(3) S. Castellino, M.R. Groseclose, and D. Wagner, Bioanalysis 3(21), 2427–2441 (2011).
(4) M.J. Kailemia, G. Xu, M. Wong, Q. Li, E. Goonatilleke, F. Leon, and C.B. Lebrilla, Anal. Chem . 90, 208–224 (2018)
(5) 3D-MASSOMICS, University of Bremen, http://eu3dmassomics.uni-bremen.de.
(6) A.D. Palme and T. Alexandrov, Anal. Chem. 87, 4055–4062 (2015).
(7) METASPACE http://metaspace2020.eu/#/about.
(8) A. Palmer, P. Phapale, I. Chernyavsky, R. Lavigne, D. Fay, A. Tarasov, V. Kovalev, J. Fuchser, S. Nikolenko, C. Pineau, M. Becker, and T. Alexandrov, Nature Methods 14, 57–60 (2017).
(9) Environmental Chemistry Methods (ECM), Analytical Methods and Procedures for Pesticides, (U.S. Environmental Protection Agency, Washington, D.C.) https://www.epa.gov/pesticide-analytical-methods/environmental-chemistry-methods-ecm-index-0-9.
(10) L. Multigner, J.R. Ndong, A. Giusti, M. Romana, H. Delacroix-Maillard, S. Cordier, B. Jégou, J.P. Thome, and P. Blanchet, J. Clin. Oncol. 28(21), 3457–3462 (2010).
(11) M. Lagarrigue, M. Becker, R. Lavigne, S-O Deininger, A. Walch, F. Aubry, D. Suckau, and C. Pineau, Mol. Cell. Proteomics 10(3), M110.005991 (2011).
(12) M. Lagarrigue, R. Lavigne, E. Tabet, V. Genet, J.P. Thomé, K. Rondel, B. Guével, L. Multigner, M. Samson, and C. Pineau, Anal. Chem. 86, 5775-5783 (2014).
(13) P. Kadhel, C Monfort, N. Costet, F. Rouget, J.P. Thomé, L. Multigner, and S. Cordier, Am. J. Epidemiol. 179(5), 536–544 (2014).
(14) Protim, https://www.protim.eu/index.php/en/.
(15) R. Lavigne et al., in preparation.
Shannon Cornett, PhD, is the Imaging Business Manager at Bruker Daltonics in Nashville, Tennessee. Mike Easterling, PhD, is the Imaging Mass Spectrometry Manager at Bruker Daltonics in Los Angeles, California. Charles Pineau, PhD, is a Research Director at Inserm and the Director of the Protim Core Facility at the University of Rennes 1, in Rennes, France. Direct correspondence about this article to Shannon.Cornett@Bruker.com

Best of the Week: AI and IoT for Pollution Monitoring, High Speed Laser MS
April 25th 2025Top articles published this week include a preview of our upcoming content series for National Space Day, a news story about air quality monitoring, and an announcement from Metrohm about their new Midwest office.
LIBS Illuminates the Hidden Health Risks of Indoor Welding and Soldering
April 23rd 2025A new dual-spectroscopy approach reveals real-time pollution threats in indoor workspaces. Chinese researchers have pioneered the use of laser-induced breakdown spectroscopy (LIBS) and aerosol mass spectrometry to uncover and monitor harmful heavy metal and dust emissions from soldering and welding in real-time. These complementary tools offer a fast, accurate means to evaluate air quality threats in industrial and indoor environments—where people spend most of their time.