CHCA or DHB? Systematic Comparison of the Two Most Commonly Used Matrices for Peptide Mass Fingerprint Analysis with MALDI MS
The authors present a comprehensive study of MALDI spotting, based upon the most commonly used dried droplet method, for various peptides of different molecular weight and concentration.
The matrix composition and the sample spotting condition have significant impact on the matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS) signal quality. In proteomics, the five most common matrices are α-cyano-4-hydroxycinnamic acid (CHCA), 2,5-dihydroxybenzoic acid (DHB), 2-(4-hydroxyphenylazo) benzoic acid (THAP), sinapinic acid (SA) and 2-(4-hydroxy-phenylazo) benzoic acid (HABA). Among these, CHCA and DHB are the most widely used for obtaining a peptide mass fingerprint. CHCA is perceived as more efficient in ionization, often resulting in higher signal intensity, and is preferred for low-abundance peptides. DHB, on the other hand, produces less background signals from matrix clusters, and thus better reveals signals in the lower m/z region and is preferred in protein posttranslational modification (PTM) studies since modifications are likely to remain intact during ionization.
In addition to the selection of matrix, the sample spotting conditions, specifically the cocrystallization of analyte with matrix, also can affect the quality of the MS signals greatly. Many reports in the literature have investigated the optimization of matrix–analyte preparation for MALDI-MS. For CHCA, various approaches have been reported for suppressing the CHCA matrix or adduct signals within the low mass region. These approaches have included the variation of CHCA concentrations (2) and the use of additives such as ammonium citrate, ammonium phosphate (2,3), nitrocellulose (4), and surfactants (5). For DHB, the focus was on sensitivity enhancement because the signal intensity obtained from DHB tends to be weaker than that obtained from CHCA (6). The use of matrix additives such as phosphoric acid (7), ammonium salt (8), and aniline (9) also has been reported for this purpose.
Direct comparisons between the performance of CHCA and DHB, as well as a mixture of CHCA and DHB, in peptide mass fingerprints have been reported by several groups. However, as summarized here, most of these studies examined a limited number of parameters. More importantly, the inconsistencies in conditions (the selection of solvents and their composition, the choice of analyte and concentration, and the use of additives) make it difficult to interpret results between different labs.
Katayama (10) first compared the effect of DHB at two different concentrations; 100 and 20 mg/mL prepared in trifluoroacetic acid–water–acetonitrile (0.1:66:33, v/v/v). Peptides from 13 silver-stained gel spots were analyzed, and the lower DHB concentration was found to produce higher signal intensities. Next, a comparison was made between DHB at 20 mg/mL and saturated CHCA prepared in the same solvent, again using peptides from 13 gel spots. In this case, DHB was found to result in comparable sensitivity to CHCA with less variation in signal intensities between peptides and better sequence coverage than CHCA. Lindahl and colleagues (11) also compared the performance of DHB and CHCA using 21 nasal lavage fluid proteins from silver-stained 2D gels. Matrix concentrations were 10 mg/mL for CHCA and 40 mg/mL for DHB, both prepared in trifluoroacetic acid–water–acetonitrile (0.3:70:30, v/v/v). Significantly more peptides with less noise and higher sequence coverage were obtained with DHB. Furthermore, peptides that were observed only with DHB were mainly within the low mass range. However, they also reported that the advantage of DHB over CHCA was not observed with peptides extracted from coomassie-stained gels.
In another report (12), the performance of CHCA and DHB were examined using five solvent systems: 99% acetone; 50% acetonitrile, 0.1% trifluoroacetic acid; 75% acetonitrile, 0.1% trifluoroacetic acid; formic acid–water–2-propanol (1:3:2); and water–methanol (2:1). Saturated solutions of both matrices were used. In-solution digests of four standard proteins were used as test analytes; however, the protein concentrations were not reported. The results did not indicate any clear advantages between these solvent systems, as all four proteins showed optimal peptide mass fingerprint performance under the different solvent systems. However, higher MOWSE (molecular weight search) scores and sequence coverages were generally obtained with DHB than with CHCA. The performance also was more consistent between the different solvent systems with DHB than with CHCA. In a report by Thiede and colleagues (6), the authors concluded that the peptide mass fingerprint signal intensities from proteins derived from the human Jurkat T-cell line on coomassie brilliant blue stained 2D gels were generally higher, as much as 17 times, with CHCA than with DHB. However, DHB was more useful in providing signals in the lower m/z region. Hence, the combination of data from CHCA and DHB resulted in higher sequence coverage than with one matrix alone. CHCA was prepared in 0.3% trifluoroacetic acid–acetonitrile (1:1) at 20 mg/mL, while DHB was prepared at 50 mg/mL in 0.3% trifluoroacetic acid–acetonitrile (2:1).
Schlosser and colleagues (13) compared the performance of CHCA, DHB, and CHCA–DHB mixture on the detection of 19 pentapeptides, each at a concentration of 0.25 mg/mL in a mixture. CHCA and DHB were prepared at 10 mg/mL in acetonitrile–water (1:1) with 0.1% trifluoroacetic acid, and the CHCA–DHB mixture was prepared by mixing these two solution at 1:1 (v/v). The use of DHB alone allowed the detection of nine peptides, while the use of CHCA alone resulted in confident detection of only one peptide. With the CHCA–DHB mixture, 17 peptides were detected, all but one as sodium adducts. The effect of matrix additives also was examined. Addition of ammonium citrate to CHCA increased the signal intensity of only one peptide but completely suppressed all others; whereas addition of potassium salt to CHCA significantly improved most peptide signals in potassium adduct forms. The use of CHCA–DHB matrix mixture also was reported by Roepstorff and colleagues (14). The analytes were peptides from low-level silver-stained 2D gel spots of 10 proteins but the quantity of proteins was not reported. CHCA at 20 mg/mL was prepared in acetonitrile–5% formic acid (7:3), and DHB at 20 mg/mL was prepared in acetonitrile–0.1% trifluoroacetic acid (7:3). The matrix mixture was prepared by mixing these at 1:1 (v/v). The matrix mixture showed increased spot-to-spot reproducibility and similar sensitivity compared to DHB, with slight improvements in sequence coverage compared to individual matrix.
As observed from this review, inconsistent conclusions have been reported in the literature due to variations in experimental conditions. To address this issue, here we present a comprehensive study on the effect of CHCA and DHB on MALDI-MS signals for peptide mass fingerprints using the dried droplet method introduced by Karas and Hillenkamp (1). A large number of parameters, including matrix solvent, composition, concentration, and the use of various additives are taken into consideration. Each of these parameters was optimized systematically for CHCA and DHB, as well as CHCA–DHB mixtures. Furthermore, to make the results more meaningful to a wide audience, a variety of peptides with different molecular weight and hydrophobicity were used as test analytes. Experiments were also performed under various analyte concentrations to reveal the effect of peptide abundance. Our intention is to provide a more comprehensive data set that can better reveal the effect on MALDI sample preparation, particularly for environments that can benefit from standardized methodologies such as high throughput analysis in proteomics facilities.
Experimental
HPLC-grade acetonitrile and ethanol (100%) were from Fisher Scientific Ltd. (Ottowa, Ontario, Canada). Fetuin was purchased from Calbiochem (San Diego, California). Other chemicals unless otherwise stated were purchased from Sigma-Aldrich (Markham, Ontario, Canada).
Sample Preparation
Bovine serum albumin (BSA), α-casein, and β-casein were digested in solution with trypsin (Promega Corp., Madison, Wisconsin). Digestion was performed in a 50 mM ammonium bicarbonate with an enzyme-to-protein ratio 1:100 (w/w). α-Casein and β-casein (bovine milk), catalase (bovine liver), apo-transferrin (bovine), α-amylase (bacillus lichenifermis), myoglobin (horse heart), BSA, cytochrome c (horse), carbonic anhydrase isozyme II (bovine erythrocytes), lysozyme (chicken egg white), ovalbumin (chicken egg), hemoglobin (human), trypsinogen (bovine pancreas), and fetuin (bovine serum) were used as protein standards. All 15 proteins (0.1 to 0.5 μg each) were separated by SDS-PAGE on 15% gels and the protein bands were visualized by colloidal coomassie staining (Gelcode Blue, PIERCE, Rockford, Illinois). In-gel reduction, alkylation, tryptic digestion and peptide extraction were performed on Waters MassPREP automated station (Milford, Massachusetts). The samples were dried in a lyophilizer and then stored at –70 °C before use. Prior to analysis, peptides were redissolved in 10% acetonitrile and 0.1% trifluoroacetic acid. DHB (98%, Aldrich 14,935-7) and CHCA (99%, Aldrich 47,687-0) were used as received. Each sample was mixed at 1:1 with matrix solution and deposited in triplicate onto the MALDI target. The spots were left to dry at ambient condition.
MALDI-TOF Mass Spectral Analysis
MS data were acquired using a MALDI TOF-TOF mass spectrometer (4700 Proteomics Analyzer, Applied Biosystems, Foster City, California) 4000 Series Explorer was used for data acquisition (Applied Biosystems, Version 3.6). The instrument is equipped with the Nd:YAG laser at 355 nm. We acquired MS spectra in the reflectron and positive ion mode by randomly irradiating the sample spot. The laser intensity was optimized for each matrix to obtain the best resolution and signal-to-noise ratios (S/N) for the peptide peaks observed. Generally, higher laser energy (by approximately 25%) was used for samples mixed with the DHB matrix compared to samples mixed with the CHCA. Data Explorer software (Applied Biosystems, version 4.6) was used to determine the S/N and relative intensity of peaks. The peak detection S/N threshold was set at 20. Relative intensity (RI) was obtained by dividing the relative intensity of each peak by that of the highest peak in the selected peptide data set. The average S/N from triplicate runs were calculated and used in all figures (Figures 1–5). The uncertainties also were determined in each case. They were shown in Figures 4 and 5 but were omitted in Figures 1–3 for visual clarity. The typical relative standard deviation ranged between 20 and 30% in Figures 1–3. All spectra were processed and analyzed by GPS Explorer software (version 3.6, ABI) with MASCOT version 2.0. Protein identification was obtained by searching the Swiss-Prot database. Protein MASCOT Scores and the number of matched peptides were used to compare CHCA and DHB performance on peptide mass fingerprint for different sample quantities.
Results and Discussion
Optimization of CHCA Matrix Preparation
CHCA has been used widely as a MALDI matrix for peptide mass fingerprint due to its high sensitivity (15). Here, CHCA matrix composition, including the concentration of CHCA, acetonitrile, and ammonium salts, ammonium citrate and ammonium phosphate, were independently optimized. Peptides from the in-solution tryptic digestion of BSA with concentrations ranging from 12.5 to 50 fmol/μL and 100 fmol/μL β-casein were used.
Acetonitrile Concentration in CHCA
The most commonly reported solvent for CHCA is 50–70% acetonitrile and 0.1% trifluoroacetic acid (11,12,14,16), most likely because of the high CHCA solubility in acetonitrile. CHCA solutions were prepared at 5 mg/mL with varying acetonitrile content (in 0.1% trifluoroacetic acid in water) from 40% to 70% at 5% intervals. Figure 1 shows the S/N ratios of six peptides from BSA with prominent signals, as well as an intense background system peak that was consistently observed at m/z of 637.37.
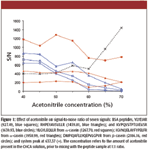
Figure 1
In general, the peptide S/N went down with increasing percentages of acetonitrile. The effect appeared to be more significant for the four hydrophilic peptides (m/z 927.5, 1439.8, 1639.9, and 1267.70). The effect of acetonitrile was less pronounced at concentrations below 50% on the relatively hydrophobic casein peptides (m/z 1958.99 and 2186.16). Finally, the intensity of the system peak at 637.37 increased significantly at acetonitrile concentrations above 60%. Based upon these observations, acetonitrile concentration of 40–50% appeared to be optimal. Considering that real samples can contain both hydrophilic and hydrophobic peptides, 50% acetonitrile was chosen to promote solubility of hydrophobic peptides. After mixing with samples at 1:1, this corresponded to a final acetonitrile concentration of 25%.
Ammonium Salt Concentration in CHCA
The addition of ammonium salts to CHCA matrix has been reported to reduce matrix cluster signals, predominantly in the mass range of 650–1300 Da, thus enhancing peptide ionization efficiency and signal intensity (2). The effect of ammonium citrate, 1–4 mM, and ammonium phosphate, 2–8 mM, were investigated as matrix additives. CHCA, 5 mg/mL, was prepared in 50% acetonitrile and 0.1% trifluoroacetic acid.
The S/N of four CHCA cluster signals and an unknown system peak at m/z 659.30 are shown in Figure 2a in the presence of different concentrations of ammonium citrate and ammonium phosphate. The assignment of these cluster signals was done by matching the m/z with that reported in the literature, and the mass errors were 0.03 Da or less (2,3). The results clearly indicate that the presence of ammonium citrate and ammonium phosphate significantly reduced the formation of matrix clusters. Their effects were however minimal on the system peak at m/z 659.30, which likely was not directly related to CHCA. Another system peak at m/z 637.3 (data not shown) also was observed consistently and could not be suppressed by the addition of ammonium citrate and ammonium phosphate. To exclude these system peaks from the MS spectra, a mass range of m/z 660 to 3500 was selected for analyses with CHCA.
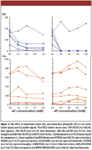
Figure 2
The S/N of seven BSA peptide peaks obtained in the presence of ammonium citrate or ammonium phosphate is presented in Figure 2b. With increasing concentration of ammonium citrate, the peptide signals either remained unchanged or decreased gradually. This observation is in agreement with a previous report (2). On the other hand, the addition of ammonium phosphate appeared to increase the peptide signals up to 6.0 mM of ammonium phosphate. This signal enhancement was particularly beneficial to three small peptides with weaker signal intensities (m/z 665.30, 712.37, and 841.46). This was also in agreement with previous report by Zhu and colleagues (2). In their work, up to 20 mM ammonium phosphate was used as CHCA additive without any negative effects on the peptide signals. Our results confirm the conclusion that ammonium phosphate is a better matrix additive compared to ammonium citrate, and that 6 mM ammonium phosphate is the optimal concentration for reducing matrix–analyte adduct formation and maximizing peptide signals. We also extended our study to include peptides from 100 fmol/μL of α- and β-casein. Similar observations in signal enhancement were obtained (data not shown), although the extent of improvement was much less than the BSA derived peptides. Later in this article, we will show that CHCA, with or without additives, is not as suitable as DHB for detecting phosphopeptides.
CHCA Concentration
The concentration of CHCA is also an important factor in determining the signal intensity of peptides. Matrix solutions containing varying CHCA concentration, from 1.25 to 10 mg/mL, were examined. All CHCA solutions were prepared in 50:50 acetonitrile and 0.1% trifluoroacetic acid in water, with 6 mM of ammonium phosphate as additive. The concentration of the peptide sample was 12.5 fmol/μL before mixing 1:1 with the matrix. The S/N values for the five BSA peptides, selected to represent signals with various intensities and m/z, are presented in Figure 3. These data clearly show that 5 mg/mL CHCA resulted in the best overall performance. At higher CHCA concentrations, much stronger matrix cluster signals were observed, which in turn suppressed the peptide signals. At lower CHCA concentrations, the matrix spot homogeneity deteriorated and it became difficult to find the "sweet" spots for good MS signals.
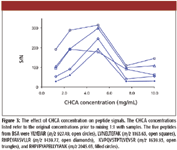
Figure 3
Optimization of DHB Matrix Preparation
DHB is another matrix widely used for peptide analysis, especially for peptides with posttranslational modification such as phosphorylation (17) and glycosylation (18), since modifications remains intact after ionization. We employed similar optimization approaches for DHB as used above for CHCA. However, in the case of DHB we began with selection of additives and solvents, followed by concentration optimization. The test analytes used in this study include peptides obtained from the in-solution digestion of 50 fmol/μL of BSA and 250 fmol/μL of α- and β-casein.
Selection of Solvent Systems and Additives for DHB Preparation
As specified in Figure 4, five solvent systems for DHB were examined for their effects on the signals of five peptides from α- and β-casein, with two of these peptides being singly phosphorylated. The use of these systems, or minor variation of these, has been reported to yield very good results (8,12,19–21). The DHB concentration was kept constant at 10 mg/mL in this part of the study. The effect of DHB concentration will be presented in the next section. The S/N values of the peptides, as well as that of the three most intense system peaks from background contaminants, were determined (shown in Figures 4a and b).
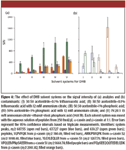
Figure 4
The results in Figure 4a suggested that the presence of additives generally increased the peptide signals. The enhancement appeared to be the most significant when both ammonium citrate and phosphoric acid with 20% ethanol were used (System V). Furthermore, the effect was observed for both phosphorylated and nonphosphorylated peptides. Although not shown in Figure 4, we also monitored the signal of a tetraphosphopeptide from β-casein (m/z 3122.4). This signal was observed only in solvents with phosphoric acid as additive (Systems III, IV, and V), and the observed S/N was the highest in Solvent V, which confirmed the importance of phosphoric acid in phosphopeptide analyses (19, 22).
The effect on system peaks was however more complex. The use of phosphoric acid seemed to be effective in suppressing the two major peaks, m/z 659.3 and 637.27, observed either with no additives or with only ammonium citrate (Systems I and II). Nevertheless, the presence of phosphoric acid introduced another prominent system peak at 607.55 (Systems III and IV). Fortunately, we were able to suppress this peak by including ethanol in Solvent System V. Together with its ability to promote the signals of both phosphorylated and non-phosphorylated peptides, System V was concluded to be the most optimal in this study.
Concentration of Solvents and Additives
In this section, we attempted to further optimize the composition of System V from the previous study by independently varying the concentration of each additive and solvent.
Phosphoric Acid: Phosphoric acid is commonly added to DHB in MALDI to acidify analytes and promote ionization, particularly for phosphopeptides (19, 22). The effect of phosphoric acid was previously studied with a concentration range of 0.01% to 10% (19). It was reported that 1% phosphoric acid resulted in high S/N for phosphopeptides without causing corrosion of the stainless steel target; however, the use of ethanol was not included in their study. Hence, we conducted our own study to vary the phosphoric acid concentration from 1 to 2% in Solvent System V. The results confirmed that 1% phosphoric acid resulted in higher S/N for both phosphopeptides and nonphosphopeptides, with increments ranging from 40 to 150% (data not shown).
DHB: The effect of varying DHB concentration was next examined. Reduction of DHB concentration from 100 to 20 mg/mL was reported to increase MALDI-MS signals of peptides (10). Because a DHB concentration of 10 mg/mL was used for the studies shown above, we subsequently examined the effect of raising its concentration to 15 and 20 mg/mL. The results indicated an overall S/N decrease at higher concentration of DHB; for example, a reduction of approximately 50% in S/N was observed at 20 mg/mL (data not shown). This confirmed that DHB concentration of 10 mg/mL was indeed the optimum.
Ethanol: In addition to acetonitrile, the use of ethanol had been reported to be suitable for the preparation of DHB (23) because DHB has a tendency to form needle-shaped crystals with acetonitrile, which makes it difficult to find the "sweet spots" for good MS signals. Accordingly, ethanol was used to re-crystalize the dried DHB-sample spot in order to improve the crystal uniformity. This led to better overall signal intensity and eliminated the need of sweet-spot searching. In a recent experiment conducted in our laboratory, we extended this idea by using ethanol (20%) as the solvent for DHB preparation in the analysis of phosphopeptides, and similar advantages were observed (21). Here, we further investigated the effect of ethanol on DHB. Matrices were prepared at varying concentration of ethanol (20, 30, 40, and 50%), while keeping all other conditions constant as Solvent System V. The S/N from four BSA peptides (m/z 698.40, 1249.67, 1889.07, and 2045.1) and a system peak at 607.55 were recorded (data not shown). The results for all four peptides improved when the ethanol content increased from 20% to 30%, and remained relatively unchanged beyond 30% ethanol. The system peak at 607.55, however also increased significantly at 30% ethanol as compared to 20%. As a result, 25% ethanol was selected for subsequent studies. It provided a marginal increase in peptide signals while keeping the system peak intensity low, which otherwise had a significant suppression effect on peptide ionization. To exclude the system peak at 607.55 from the spectra, a mass range of m/z 610 to 3500 was used for subsequent analyses with DHB.
Ammonium Citrate: The concentration of ammonium citrate was varied from 0 to 17.5 mM, and the resulting S/N of five BSA peptides is shown in Figure 5. Generally, the peptide S/N increased with increasing concentration of ammonium citrate up to 15 mM, confirming this to be the optimal value. The same experiment was repeated for peptides from α-casein and β-casein, and the same observation was obtained (data not shown). Finally, we also investigated the effect of ammonium phosphate in DHB. Readers are reminded that ammonium phosphate was shown earlier to be a better additive than ammonium citrate for CHCA matrix. Therefore, experiments were conducted with ammonium phosphate as the additive instead of ammonium citrate, at concentrations of 5, 10, and 15 mM. Based upon S/N of the same five peptides from BSA, the presence of ammonium phosphate did not show any significant improvement (data not shown).
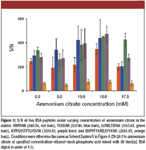
Figure 5
Comparison of CHCA and DHB at Various Sample Concentrations
As indicated previously, the results reported in the literature on the comparison of CHCA and DHB were somewhat inconsistent (10–12). These inconsistencies can arise from the different methods employed for matrix preparation, as well as the different sample concentrations used in the studies. In this section, we performed a direct comparison of the two matrices at varying sample concentration, using the matrix preparation formulations optimized previously; namely, 5 mg/mL CHCA in 50% acetonitrile with 0.1% trifluoroacetic acid and 6 mM ammonium phosphate, and 10 mg/mL DHB in 25% ethanol with 1% phosphoric acid and 15 mM ammonium citrate. The tryptic peptides from 15 protein standards were analyzed at various concentrations up to approximately 500 fmol/μL. The averaged MASCOT scores and numbers of peptides matched by the search engine, based upon triplicate measurements were listed in Table I, along with the specific sample concentration used.
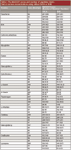
Figure 6
For most proteins, the difference between CHCA and DHB was noticeable at the highest and lowest concentrations. At higher concentrations, the use of DHB allowed detection of more peptides and thus resulted in higher scores compared to CHCA. However, the opposite was observed at low analyte concentrations, where higher number of peptides and scores were obtained with CHCA. The performances of the two matrices were more similar at the intermediate concentrations studied (20–200 fmol/μL), although DHB appeared to be marginally superior for 13 proteins. The data confirmed the common perception of CHCA of being more efficient in ionization, namely it offered higher sensitivity when the analyte concentration was low. On the other hand, the use of DHB on intermediate or high concentration samples allowed the detection of higher numbers of peptides, which could lead to a more complete protein sequence coverage. However, what the data in Table I did not tell us was whether there were any differences in properties, such as molecular weight, between the peptides identified with CHCA and those with DHB. We attempted to address this question in the following section.
Signal Bias of CHCA and DHB on Peptide Molecular Weight
The choice of CHCA or DHB had been reported to have different effects on the relative signal intensities among the peptides within a mixture (6,24), and thus the use of CHCA versus DHB can result in detection of different peptides from the same sample. The complementary information obtained from these two matrices had been combined to increase amino acid sequence coverage and the confidence of the protein identification (6,11,12). In this section, data are presented from experiments that were performed to examine this phenomenon more closely, namely to determine how the matrices have ionization bias on peptides with different molecular weight.
Peptides obtained from the in-gel digestion of 15 protein standards with a variety of characteristics were used: transferrin (32 fmol/μL), BSA (63 fmol/μL), fetuin (109 fmol/μL). trypsinogen (219 fmol/μL), catalase (52 fmol/μL), hemoglobin subunit alpha (94 fmol/μL), hemoglobin subunit beta (197 fmol/μL), α-amylase (57 fmol/μL), myoglobin (197 fmol/μL), cytochrome C (113 fmol/μL), α-casein (64 fmol/μL), β-casein (89 fmol/μL), ovalbumin (234 fmol/μL), carbonic anhydrase (215 fmol/μL), and lysozyme (308 fmol/μL). Peptide mass fingerprint analysis was performed individually on the peptides from one protein at a time, using either CHCA or DHB. Triplicate measurements were performed on each protein. The m/z range of 640 to 3000 for DHB, and 660 to 3000 for CHCA were used. All signals with S/N above 20 were recorded. The relative intensities were determined by normalizing against the strongest peak within each analysis. The peptide signals from all proteins were complied and sorted into m/z to reveal differences in MW bias between the two matrices (Figure 6). This figure is an expanded view of a table listing the peptide signals in increasing order of m/z. The approximate m/z is marked in red for CHCA and blue for DHB. The RI are divided into four ranges: 0–25%, 25–50%, 50–75%, and 75–100%; they are represented in four levels of darkness (white, light gray, dark gray, and black respectively).

Figure 7
A number of conclusions can be drawn from this figure. Using the same peptide mixtures, a higher number of total peptides, 341, were observed using CHCA at the optimized condition compared to that obtained with DHB at the optimized condition (298 total peptides). Furthermore, while not illustrated in Figure 6, the observed signal ion counts were generally higher with the CHCA matrix. Using the result from 63 fmol BSA as an example, S/N of the top 10% ranged from 380 to 1040 with CHCA, while that of DHB ranged from 253 to 499. This is again in agreement with the literature that the use of CHCA often results in better sensitivity than DHB.
More interesting remarks can be drawn by examining the m/z distribution of the identified peptides with the two matrices. A higher number of peptides at the lower m/z range (up to ~1000) were identified with DHB, and many of these smaller peptides had high RIs of 25% and above. In contrast, fewer peptides were detected at this low m/z range with CHCA, and most of them were weak in RI. However, CHCA is more efficient in producing peaks at higher m/z; for example, 1500 and above in this case. This confirmed the complementary nature of the two matrices that has been reported in the literature. The presence of the small peptides is valuable in improving the quality of the peptide mass fingerprint search results; on the other hand, large peptides are useful in producing more fragment ions in MS-MS. For this reason, complementary information can be obtained by performing peptide mass fingerprint analysis using both CHCA and DHB.
CHCA–DHB Mixture as a Co-Matrix
The use of a CHCA–DHB mixture as MALDI matrix was reported previously to produce better sensitivity and improve ionization in peptide mass fingerprint analysis (13,14). Specifically, the CHCA–DHB mixture composition according to reference 14 was 10 mg/mL CHCA and 10 mg/mL DHB dissolved in 60% acetonitrile containing 2.5% formic acid and 0.05% trifluoroacetic acid. In recognition of the additive and solvent optimization results presented earlier, we proposed another formulation of CHCA–DHB mixture using a 1:1 mixture of the two matrix solutions at the optimized conditions. Once again, our optimized CHCA solution was 5 mg/mL CHCA in 50% acetonitrile with 0.1% trifluoroacetic acid and 6 mM ammonium phosphate, and our optimized DHB solution was 10 mg/mL DHB in 25% ethanol with 1% phosphoric acid and 15 mM ammonium citrate. Experiments were conducted to evaluate the performance of our CHCA–DHB co-matrix, in comparison to the formulation used in reference 14, using test peptides from the tryptic digestion of 30 fmol/μL BSA and a mixture of 75 fmol/μL α- and β-caseins.
The data, not presented in detail, are summarized as follows. Our co-matrix of the 1:1 mixture appeared to be more effective in suppressing the CHCA matrix cluster signal compared to that prepared according to reference 14. For example, a strong signal of CHCA adduct was observed at m/z 877.05 in the co-matrix by reference 14, but not in our CHCA–DHB. However, the suppression with this co-matrix was not as good as that observed in Figure 2a, in which the single CHCA matrix was prepared at the optimized condition (with ammonium phosphate as additive). In terms of peptide signals, between the two co-matrices, our 1:1 mixture also resulted in higher overall S/N. For example, a 10- and 2.5-fold difference was observed respectively for a BSA peptide (m/z 689.3) and a phosphopeptide from β-casein (m/z 2016.82). Nevertheless, the peptide signals of the co-matrix generally were weaker than that of CHCA alone at the optimized condition. For instance, BSA peptides, MPCamTEDYLSLILNR (m/z 1724.84) and RHPYFYAPELLYYANK (m/z 2045.03), which were clearly detected with CHCA, were not detected with the two co-matrices. When compared to DHB, the co-matrix was not superior either. Some small peptides (such as m/z 742.44) detected at significantly lower S/N, and some phosphopeptides (m/z 1466.60 and 1927.68) were not detected when the CHCA–DHB mixtures were used. In summary, the combined CHCA–DHB behaved somewhat as an intermediate of the two individual matrices, and did not result in any unique advantages over each of the individual matrices CHCA and DHB at optimized conditions.
Conclusion
The optimization of additive and solvent composition of both CHCA and DHB matrices significantly improved the quality of the MALDI-MS data in peptide mass fingerprint analysis. Our results indicated a differential bias between the two matrices toward peptides of different molecular weight and at different peptide concentration. In general, for peptides extracted from a dark gel spot, or protein concentration of high fmol-level or above, the use of DHB should result in identification of higher numbers of peptides and better database search scores. Therefore, under these conditions, DHB should be considered as the preferred matrix. By comparison, in the case of weakly visible gel spots, or low-femtomole level of proteins, the use of CHCA offered better sensitivity and the detection of more peptides. However, because it is important to recognize that peptide mass fingerprint results can vary significantly from protein to protein, these recommendations might not apply universally for all proteomics studies. Finally, based upon the complementary peptide bias of CHCA and DHB, the most comprehensive information can be obtained by performing two analyses with each of the two matrices.
Acknowledgments
The authors would like to acknowledge the financial support of the Schulich School of Medicine & Dentistry and the Department of Biochemistry of the University of Western Ontario, Canada. The mass spectrometer was funded by the Academic Development Fund Program of UWO. Financial support for C.J. was provided by the Canada Foundation for Innovation through the Infrastructure Operating Funds. Services provided by the UWO Functional Proteomics and MALDI-MS Facilities also are acknowledged.
Cunjie Zhang, Haixia Zhang, David W. Litchfield, and Ken K.-C. Yeung are with the Functional Proteomics and MALDI-MS Facilities, Department of Biochemistry, Schulich School of Medicine and Dentistry, University of Western Ontario, Canada.
References
(1) M. Karas and F. HillenKamp, Anal. Chem. 60, 2299–2301 (1988).
(2) X. Zhu and I.A. Papayannopoulos, J. Biomolecular Techn. 14, 298–307 (2003).
(3) I.P. Smirnov et al., Anal. Chem 76, 2958–2965 (2004).
(4) M. Donegan, A.J. Tomlinson, H. Nair, and P. Juhasz, Rapid Commun. Mass Spectrom. 18, 1885–1888 (2004).
(5) D.C. Grant and R.J. Helleur, Rapid Commun. Mass Spectrom. 21, 837–845 (2007).
(6) B. Thiede, F. Siejak, C. Dimmler, P.R. Jungblut, and T. Rudel, Electrophoresis 21, 2713–2720 (2000).
(7) H.-C. Hsieh, C. Sheu, F.-K. Shi, and D.-T. Li, J. Chromatogr., A 1165, 128–135 (2007).
(8) J.M Asara, and J. Allison, J. ASMS 10, 35–44 (1999).
(9) S.I. Snovida, V.C. Chen, and H. Perreault, Anal. Chem. 78, 8561–8568 (2006).
(10) H. Katayama, T. Nagasu, and Y. Oda, Rapid Commun. Mass Spectrom. 15, 1416–1421 (2001).
(11) B. Ghafouri et al., Anal. Biochem. 371, 121–123 (2007).
(12) N. Padliya and T.D. Wood, Proteomics 4, 466–473 (2004).
(13) G. Schlosser, G. Pocsfalvi, E. Huszar, A. Malorni, and F. Hudecz, J. Mass Spectrom. 40, 1590–1594 (2005).
(14) S. Laugesen and P. Roepstorff, J. ASMS 14, 992–1002 (2003).
(15) R.M. Whittal, B.O. Keller, and L. Li, Anal. Chem. 70, 5344–5347 (1998).
(16) S.J. Cordwell et al., Proteomics 8, 122–139 (2008).
(17) M.R. Larsen, T.E. Thingholm, O.N. Jensen, P. Roepstorff, and T.J.D. Jorgensen, Molecular Cellular Proteomics 4, 873–886 (2005).
(18) S.I. Snovida and H. Perreault, Rapid Commun. Mass Spectrom. 21, 3711–3715 (2007).
(19) S. Kjellstrom and O.N. Jensen, Anal. Chem. 76, 5109–5117 (2004).
(20) H. Zhang, C. Zhang, G.A. Lajoie, and K.K.C. Yeung, Anal. Chem. 77, 6078–6084 (2005).
(21) H. Zhang, G.K. Hunter, H.A. Goldberg, G.A. Lajoie, and K.K.C. Yeung, Anal. Chim. Acta 581, 268–280 (2007).
(22) A. Tholey, Rapid Commun. Mass Spectrom. 20, 1761–1768 (2006).
(23) D.J. Harvey et al., Organic Mass Spectrom. 29, 753–765 (1994).
(24) J. Gobom, E. Nordhoff, E. Mirgorodskaya, R. Ekman, and P. Roepstorff, J. Mass Spectrom. 34, 105–116 (1999).

Smarter Sensors, Cleaner Earth Using AI and IoT for Pollution Monitoring
April 22nd 2025A global research team has detailed how smart sensors, artificial intelligence (AI), machine learning, and Internet of Things (IoT) technologies are transforming the detection and management of environmental pollutants. Their comprehensive review highlights how spectroscopy and sensor networks are now key tools in real-time pollution tracking.