Looking Below the Surface of Breast Tissue During Surgery
Special Issues
In this article, we present a method that provides prompt detection of the presence of cancer cells inside the 2-mm margin of tissue surrounding the tumor after excision using spatially offset Raman spectroscopy (SORS). SORS was developed to detect subtle changes in soft tissue spectra in the 100–2000 ?m range and tested on excised breast tissues. The results display a very high specificity and sensitivity (100% and 95%, respectively) of classification between positive and negative tumor margins. SORS is a clinically feasible method, suitable for the real-time, intraoperative assessment of tumor margins at the micrometer level.
In this article, we present a method that provides prompt detection of the presence of cancer cells inside the 2-mm margin of tissue surrounding the tumor after excision using spatially offset Raman spectroscopy (SORS). SORS was developed to detect subtle changes in soft tissue spectra in the 100–2000 μm range and tested on excised breast tissues. The results display a very high specificity and sensitivity (100% and 95%, respectively) of classification between positive and negative tumor margins. SORS is a clinically feasible method, suitable for the real-time, intraoperative assessment of tumor margins at the micrometer level.
One in eight women is diagnosed with breast cancer today, making it the most frequently diagnosed cancer among women (1). Breast cancer is the second leading cause of cancer-related deaths among women in the United States; however, when diagnosed early, patients with localized breast cancer have a 98% survival rate. Treatment relies on surgery for the complete management of patient care. Of the patients who are treated by surgery, 75% are eligible for breast conserving therapy (BCT), also known as a partial mastectomy or lumpectomy. In this procedure, tumors are first reduced using neoadjuvant therapies and then excised locally to minimize the cosmetic and psychological impact on the patient. This surgery is less invasive than a full mastectomy and has been shown to provide similar long-term survival rates (2).
Recurrence is directly related to the complete removal of all remnant tumor cells. Thus, the typical clinical standard for a successful partial resection is to have a 1–2 mm negative (tumor-free) margin around the tumor in the excised tissue. Margins therefore play a key role in the prognosis of the patient with respect to local recurrence of breast cancer and are directly correlated to the success of BCT as a treatment modality. This then leads to a critical need for intraoperative evaluation of the resection front so that immediate re-excision of suspicious margins can be performed, minimizing the necessity for a second surgery.
Any method used to evaluate the surgical margins must be rapid and relatively simple to implement if it is to be used in routine clinical care. The simplest technique for determining margin status is visual inspection of the excised tissue for evidence of tumor, a method that leads to incorrect diagnoses, and therefore repeat surgeries or a higher risk of recurrence in at least 25% of cases (3). Serial sectioning with standard histopathology provides a definitive diagnosis of margin status, but results may take several days to over a week. This delay leaves a patient uncertain and waiting and the results may then lead to a second surgery if tumor-positive margins are found (20–70% of cases). Limitations in current methods therefore emphasize the need for a real-time, intraoperative tool that can accurately determine the status of breast surgical margins.
In this article, we present the development of spatially offset Raman spectroscopy (SORS) for subsurface assessment of surgical margins in the spatial scale of 100–2000 μm. The scientific goal of the project is to evaluate the ability of SORS to perform biochemical separation of subtly differing tissues in the needed spatial scale such that margin assessment can be performed. The clinical goal of the study is to develop a method to detect the presence of tumor cells within the clinically accepted margin area of 1–2 mm in real time, in the operating room so that the surgeon is provided with real-time feedback and additional tissues may be removed in a single surgical procedure to achieve clear margins and eliminate the need for a second surgery.
Optical Methods and Margin Analysis
Raman spectroscopy is based on inelastic scattering involving transitions between vibrational energy levels. A Raman spectrum therefore consists of a series of peaks, which represent the different vibrational modes of the scattering molecules. These peaks are spectrally narrow and molecular-specific, such that the observed peaks may be associated with specific bonds in specific molecules. Many biological molecules have distinct Raman spectra that can be used to determine the biochemical composition of a tissue. One particularly relevant biochemical change in cancer cells is an increase in the nucleic acid content that may be correlated with increased proliferation and genetic instability. This change, among others such as changes in glycogen and collagen, can be detected with Raman spectroscopy and used for cancer detection (4,5).
Many researchers have exploited such cancer-specific signatures with Raman spectroscopy in many organs, including the cervix (6,7), bladder and prostate (8), lung (9), skin (10,11), and the gastrointestinal tract (12–14). Several groups have addressed the application of Raman spectroscopy for breast cancer diagnosis. Alfano and colleagues (15–18) were the first to look at Raman spectroscopy to distinguish normal from malignant breast tissues. More recently, Feld and colleagues (19) performed extensive work on using Raman spectroscopy for breast cancer diagnosis and can discriminate malignant from normal and benign tissues with 94% sensitivity and 96% specificity. It should be noted that all the work described above focuses on breast cancer diagnosis and not on guidance of therapy or margin assessment.
Beyond the use of Raman spectroscopy, there have been some reports of novel methods for margin assessment, including the use of radiofrequency spectroscopy (20), elastic scattering spectroscopy (21), optical coherence tomography (OCT) (22), and diffuse reflectance spectroscopy (23); however, most of these reports suffer from poor sensitivity or specificity. OCT can provide high-resolution images that can be used for guidance in margin analysis. Recently, Boppart and colleagues (22) showed that OCT performs with an overall accuracy of 75% for determining positive tumor margins. Ramanujam and colleagues (23) have reported using diffuse reflectance spectroscopy for margin assessment achieving an overall accuracy again of 75%. These studies provide improvements in comparison to current clinical standards for intraoperative detection; however, they are limited in their impact on patient care.
One report on breast margin analysis with Raman spectroscopy was published by Feld and colleagues (24), where normal, fibrocystic, and malignant tissues were classified with an overall accuracy of 93%. However, this report relies on a standard fiber probe configuration that collects signal from only the first several hundred micrometers in depth and does not consider the need for determining negative margins to a depth of 1–2 mm. Whereas Raman spectroscopy can provide specific biochemical information for breast tumor margin analysis, the ideal optical system would be able to resolve this information at depths of at least 2 mm and do so in seconds to minutes in the operating room.
Spatially Offset Raman Spectroscopy
Optical techniques rely on the penetration of light to obtain depth-related or resolved information. This in turn depends on the wavelength of light under use. Introducing a spatial offset between source and detection elements provides the depth selectivity in spectral measurements needed for this application. Figure 1 demonstrates the photon walk that provides this depth resolution. Larger separations are more likely to detect photons that have traveled deeper into the tissue and have been scattered multiple times compared with smaller separations, which detect photons that have only undergone minimal scattering events and have remained in the superficial layers. Matousek and colleagues first demonstrated this effect by obtaining Raman spectra of diffusely scattering media using a two-layer phantom in which each layer had a chemical with a distinctive spectrum. As the source–detector offset increased, the measured spectra began to resemble a pure spectrum from the bottom layer (25). The same group demonstrated the first biological application of this technique in detecting the strong Raman signature of bone through several millimeters of soft tissue (26). However, the application of SORS was limited to detecting a small area of very strong scatterers, such as bone under soft tissue. The results presented in this article demonstrate the application of SORS to discriminate multiple layers of soft tissue, and more particularly, for detecting cancer through underlying layers of normal tissue on the submillimeter scale.
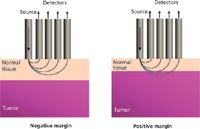
Figure 1: Schematic illustrating tumor-positive and tumor-negative margins, defined by the distance between the surgical margin and the tumor boundary. Overlaid are possible photon migration paths demonstrating the advantage of SORS for this application.
The fundamental principle of SORS therefore could prove to be very useful for obtaining measurements from a number of tissue depths up to the clinically relevant 1–2 mm in resected partial mastectomy specimens. To demonstrate detection of subtly different soft tissues in depth, a tissue phantom model consisting of a layer of normal human breast tissue was placed between two 100-μm-thick quartz coverslips. The normal layer thickness was varied from 0.5 to 2 mm and placed directly on top of breast cancer tissue sample, which ranged in thickness from 2 to 5 mm. With one excitation fiber (S) to deliver the laser and one fiber (D) to collect the Raman signal, measurements were taken by varying the distance between the source and detector (S, D) fibers ranging from 0.75 to 4.75 mm in 0.5 mm intervals. Figure 2 shows spectra obtained from the experimental setup with 1-mm normal layer thickness, as well as the mean spectrum of normal and tumor layers (measured directly). As spatial offset increases, the spectra begin to resemble the tumor spectrum compared to the normal spectrum.
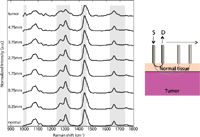
Figure 2: Raman spectra from an experimental run with a 1-mm normal layer. Gray boxes highlight regions with the most dramatic changes from normal to tumor signatures as S-D offset (labeled on left) increases.
The light gray boxes in Figure 2 highlight the spectral regions with the most dramatic changes observed as spatial offset increases. A close-up of these regions is shown in Figure 3. These include the increase of the 1006-cm-1 peak generally attributed to phenylalanine, a ratio of the 1303–1265 cm-1 peaks, which tends to indicate an increase in protein content, and the increasing width of the amide I peak around 1656 cm-1. Other significant changes are a decrease in the relative intensity of the 1445-cm-1 CH2 deformation peak, a decrease in the 1748-cm-1 carbonyl stretch peak, and an increase in the 1156-cm-1 carotenoid peak. These results show that SORS can be used to detect spectral contributions from breast tumors under normal breast tissue.

Figure 3: To aid in the visualization of relevant but subtle spectral changes, zoomed-in versions are shown for the 1006-cm-1 phenylalanine peak, the 1265-cm-1 amide III and 1303-cm-1 lipid peak, the CH2 bend/deformation peak at 1440 cm-1, and the shoulders of the 1656-cm-1 amide I peak.
To translate this research feasibility into a clinically applicable setup, a fiber-based probe was designed to obtain a sampling depth of 2 mm so that the probe is sensitive to the presence of tumor cells within this depth. Additionally, the source–detector offset was fixed so that only Raman signal from the superficial 2-mm layers of tissue was collected. Based on experimental (27) and computer-simulated results (28), a maximum source–detector offset of 3.5 mm was been determined to provide the collection geometry needed to detect the presence of breast tumors located up to 2 mm beneath normal breast tissue. If a larger offset is used, the measurements could detect tumors from more than 2 mm deep as needed.
A multiple offset SORS probe was constructed (assembled by EMVision, Loxahatchee, Florida), consisting of a single 400-μm-diameter source fiber on one end, with four (partial) rings of 300-μm-diameter collection fibers (numbered R1, R2, R3, and R4). Inline filters were placed to reject elastically scattered light generated in the fibers. As in a typical clinical Raman setup, the source probe delivered 80 mW of power from a 785-nm diode laser (Innovative Photonics Solutions, Monmouth Junction, New Jersey). The detection fibers delivered light to a near-IR-optimized spectrograph (Princeton Instruments, Princeton, New Jersey), which dispersed the light to be recorded by a deep depletion, thermoelectrically cooled CCD (Princeton Instruments). A 70-μm entrance slit is used and the spectral resolution achieved by the system is about 11 cm-1 in the 900–1800 cm-1 spectral region. Spectra were calibrated for instrument variation and system response and processed for noise and fluorescence background (26).
SORS for Breast Tumor Samples
With approval from the Vanderbilt Institutional Review Board (Vanderbilt University, Nashville, Tennessee) and the US Army Medical Research and Materiel Command's Human Research Protection Office, 35 human breast tissue samples were obtained from women undergoing partial mastectomies or full mastectomies. Of those samples, 15 had areas greater than 2 mm of normal tissue with no tumor; the remaining 20 samples had positive margins, defined by less than 2 mm of benign tissue above tumor. To acquire Raman spectra from samples with positive margins, the SORS probe was placed on an area of normal-appearing tissue above the tumor to replicate the standard clinical protocol for tumor margin evaluation. Spectra were recorded for 10–30 s and processed. To compare the spectra to the histopathological evaluation of margin assessment, measurement sites were inked, fixed in formalin, and serially sectioned.
Two tissue-margin scenarios and their resulting spectra are displayed in Figure 4. The "S" arrow designates the spot of the excitation fiber and R1–R4 indicate the placement of the detection fiber rings. Figure 4a shows the spectra obtained from the tissue section shown in the insert. In this example, the source fiber delivered light to a large fatty region, the area appearing white under H&E stain. The outermost collection fiber (R4) gathered photons that passed through a large tumor section, the darker pink area. Although the spectral changes are subtle, measurements acquired from the rings closer to the tumor were more similar to pure tumor spectra, suggesting that each ring was obtaining information from a different volume of the sample. These changes occur as the source–detector offset increases and are similar to those seen in Figure 2.

Figure 4: (a) SORS spectra for each detector ring from tissue in inset with a large area of normal fat (white colored area labeled "N") on the right, and solid tumor (darkly stained area labeled "T") on the left. Arrows indicate the placement of the source fiber (S) and each of the detector rings. (b) SORS spectra for each detector ring from tissue in inset with pockets of normal fat near surface of otherwise darkly stained tumor tissue. Arrows again represent placements of fibers.
Figure 4b demonstrates a case where a clear delineation between normal and tumor does not exist. The source fiber of the probe was placed on the small and narrow benign area near the surface and detection fibers were mostly on the area of the tumor. Accordingly, the spectra obtained from this tissue sample contain features indicative of normal fatty breast tissue from the fiber with the smallest source–detector offset (that is, closest to the excitation source), as well as pure tumor from fibers with larger source–detector offsets. Thus, it is clear that different detector rings are sampling multiple volumes as desired and that the SORS probe can collect data showing biochemical differences at varying depths.
Clinically, it is only necessary to determine if a given measurement is positive for the presence of tumor cells in the 2-mm probe depth of the tissue. To implement this clinically relevant scheme, spectra from all the rings were averaged to create one spectrum per site measured. Discrimination was performed with sparse multinomial logistic regression (SMLR) (29), a Bayesian machine-learning framework that computes the posterior probability of a spectrum belonging to a tissue class based on a training set. In the case of this binary analysis (either positive or negative margins), whichever class had the higher probability of membership is where the spectrum classified. Table I shows the confusion matrix for classification of these composite spectra using SMLR. These results show that SORS performs at 95% sensitivity and 100% specificity in evaluating breast tumor margins (30).
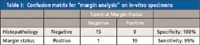
Table I: Confusion matrix for "margin analysis" on in-vitro specimens
With the promising results from this in-vitro study, patients with breast tumors were recruited to a pilot clinical study, also approved by the Vanderbilt IRB, to determine if SORS can be used to detect positive and negative tumor margins in the operating room on ex-vivo excised specimens during surgery. To standardize the study protocol in using the current SORS probe that measures a single 3.5-mm area at a time, one location was measured from each of the six surfaces of a cuboid resection specimen, as indicated in the insert of Figure 5. These sites were subsequently inked for histological correlation. Figure 5 shows the composite spectra acquired from each of these six sites in a given specimen. SORS spectra indicate the presence of tumor cells within the 2-mm margin in locations 2, 4, and 6, which was confirmed histologically (data not shown). Locations 1, 3, and 5, however, show no cancer within the margins. This example demonstrates the power of SORS for margin analysis in a clinical setting. However, simultaneous or sequential measurement of the entire tissue surface will be required for this technique to become standard clinical practice in the operating room. This work is currently in progress in our laboratory.

Figure 5: Spectra from six different anatomical positions (from 1 to 6) on an excised lumpectomy from a patient.
Conclusions and Future Considerations
This article demonstrates that SORS can be used to detect the spectral signatures of breast tumors as small as 1–2 mm thick under up to 2 mm of normal breast tissue. This study also shows that SORS can provide resolution on the order of micrometers and can detect layers with subtle differences in biochemical patterns. Being able to collect data from such small areas deeply embedded within samples paves the way for SORS to be used for many biomedical and other applications. For example, when treating patients with severe burns and wounds, it is difficult for medical providers to detect the stage of healing or the presence of infection. SORS could be used for this purpose so doctors could noninvasively detect changes below the surface. Acquiring Raman spectra from various depths in tissue may also make other cancers such as skin, cervical, and colon easier to diagnose. Many nonbiological applications may also benefit from SORS. The Drug Enforcement Agency and Homeland Security could implement SORS screening of unknown substances to obtain vital information from multiple layers and determine if any illegal substances or sources of chemical warfare are present. These are some of the potential uses of SORS. From our intraoperative tumor detection applications to the work in pharmaceutical applications, SORS has opened the door to looking beyond the surface.
Acknowledgments
The authors would like to acknowledge the financial support of the Department of Defense Breast Cancer Research Program Idea Award #W81XWH-09-1-0037.
References
(1) Cancer Facts & Figures 2009. American Cancer Society (2009).
(2) Breast Cancer Facts & Figures 2009–2010, in American Cancer Society, American Cancer Society, Inc.: Atlanta (2010).
(3) G.C. Balch et al., Am. Surg. 71(1), 22–7; 27–8 (2005).
(4) A. Mahadevan-Jansen, Raman Spectroscopy: From Benchtop to Bedside, in Biomedical Photonics Handbook, T. Vo-Dinh, Ed. (CRC Press, Washington, DC, 2003), pp. 30:1-30:27.
(5) A. Mahadevan-Jansen and R. Richards-Kortum, J. Biomed Optics 1, 31–70 (1996).
(6) E.M. Kanter et al., J. Biophotonics, 2(1-2), 81–90 (2009).
(7) E.M. Kanter et al., Am. J. Obstet. Gynecol. 200(5), 512 e1–5 (2009).
(8) P. Crow et al., Urology 65(6), 1126–1130 (2005).
(9) Z. Huang et al., Int. J. Cancer 107(6), 1047–1052 (2003).
(10) C. Lieber et al., Lasers Surg. Med., 2008 (in press).
(11) S. Sigurdsson et al., IEEE Trans. Biomed. Eng. 51(10), 1784–1793 (2004).
(12) G. Shetty et al., Br. J. Cancer 94(10), 1460–1464 (2006).
(13) M.G. Shim et al., Photochem. Photobiol. 72(1), 146–150 (2000).
(14) A. Molckovsky et al., Gastrointest. Endosc. 57(3), 396–402 (2003).
(15) R.R. Alfano et al., J. Opt. Soc. Am. B 6, 1015–1023 (1989).
(16) R.R. Alfano, Laser Life Sci. 4, 23–28 (1991).
(17) G.C. Tang, A. Pradhan, and R.R. Alfano, Lasers Surg. Med. 9(3), 290–295 (1989).
(18) C.H. Liu et al., Journal of Photochemistry and Photobiology B-Biology 16(2), 187–209 (1992).
(19) A.S. Haka et al., Proc. Natl. Acad. Sci. USA 102(35), 12371–12376 (2005).
(20) T. Karni et al., Am. J. Surg. 194(4), 467–473 (2007).
(21) I.J. Bigio et al., J. Biomed. Opt. 5(2), 221–8 (2000).
(22) F.T. Nguyen et al., Cancer Research 69(22), 8790–8796 (2009).
(23) J.Q. Brown et al., IEEE Journal of Selected Topics in Quantum Electronics: A Publication of the IEEE Lasers and Electro-optics Society 16(3), 530–544 (2010).
(24) A.S. Haka et al., Cancer Res. 66(6), 3317–3322 (2006).
(25) P. Matousek et al., Appl. Spectrosc. 59(4), 393–400 (2005).
(26) P. Matousek et al., Appl. Spectrosc. 60(7), 758–763 (2006).
(27) M.D. Keller, S.K. Majumder, and A. Mahadevan-Jansen, Opt. Lett. 34(7), 926–928 (2009).
(28) M.D. Keller, R.H. Wilson, M.A. Mycek, and A. Mahadevan-Jansen, Applied Spectroscopy, 64(6), 607–614 (2010).
(29) B. Krishnapuram, IEEE Transactions on Pattern Analysis and Machine Intelligence 27(6), 957–968 (2005).
(30) M.D. Keller, E. Vargis, N.M. Granja, R.H. Wilson, M.A. Mycek, M.C. Kelley, and A. Mahadevan-Jansen, Journal of Biomedical Optics, in review (2011).
Anita Mahadevan-Jansen, Elizabeth Vargis, Brittany Caldwell, and The-Quyen Nguyen are with the Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee.
Matthew D. Keller is with Lockheed Martin Aculight Corporation, Bothell, Washington.
Nara de Matos Granja and Melinda Sanders are with the Division of Pathology at Vanderbilt University Medical Center, Nashville, Tennessee.
Mark C. Kelley is with the Division of Surgical Oncology at Vanderbilt University Medical Center, Nashville, Tennessee.
New Study Reveals Insights into Phenol’s Behavior in Ice
April 16th 2025A new study published in Spectrochimica Acta Part A by Dominik Heger and colleagues at Masaryk University reveals that phenol's photophysical properties change significantly when frozen, potentially enabling its breakdown by sunlight in icy environments.
