Microfluidic Chip-Based Technology as a Growing Trend in LC-MS Analysis
This article describes the use of a microfluidic chip system, paired with a simple, automated interface to a wide range of mass spectrometers for both protein and small molecule applications.
The ever-increasing demands on liquid chromatography–mass spectrometry to provide higher sensitivity, accuracy, and precision with even smaller sample sizes, for a wide variety of applications, have challenged the limits of the capabilities of conventional approaches. This article describes the use of a microfluidic chip system, paired with a simple, automated interface to a wide range of mass spectrometers for both protein and small molecule applications.
Mass spectrometry (MS) has been the method of choice for highly sensitive and specific analysis of small molecules for many years, and its utility for this application continues in areas such as the testing of food for contaminants. Over the past two decades, the most significant new application for MS has been the analysis of proteins. Much of our current understanding of proteomics is derived from MS studies of protein identification, changes in protein expression, and posttranslational modifications (PTMs) in a wide range of organisms.
Electrospray ionization (ESI) and matrix-assisted laser desorption–ionization (MALDI) are the two techniques most commonly used to produce protein and peptide ions for MS analysis. MALDI, in combination with time-of-flight (TOF) MS, often is used for protein analysis due to its mass accuracy and speed of analysis. ESI can be integrated with either ion-trap, triple-quadrupole (QQQ), or quadrupole-TOF (Q-TOF) MS to provide protein and peptide identification, quantitation, and posttranslational modification analysis.
The need for sensitivity has driven the development of very-small-bore high performance liquid chromatography (HPLC) columns with nanospray interfaces to MS. The sensitivity of detection is proportional to the square of the column diameter, so small-bore columns can provide significant improvements in limits of detection. However, the use of such nanoflow LC systems with a nanospray MS interface necessitates the use of small capillary tubing connections, which can result in frequent clogging, leaking, or poor spray.
HPLC and Electrospray on a Chip
The need to provide easy-to-use, highly reliable, and highly sensitive LC–MS systems has led to the development of a microfluidic chip that integrates all of the necessary components directly onto a reusable biocompatible polymer chip (1,2). Solvent and sample delivery to the chip, high-pressure switching of flows, and automated chip loading and positioning in the MS source are accomplished by an HPLC-chip–MS interface module. The reusable microfluidic chip integrates the sample enrichment and separation columns of a nanoflow LC system with microvalve connections and the nanospray tip for interfacing with a mass spectrometer, directly on a biocompatible polyimide chip (Figure 1).

Figure 1
The HPLC chip system described here is fabricated from polyimide film that is resistant to most solvents, tolerates a wide pH range, and is compatible with the analysis of peptides, proteins, and organic molecules. The fabrication process uses laser ablation in combination with vacuum lamination of the film to create a multilayer microfluidic device (1). The resulting microchannels are packed with column materials, and metals are applied by thin-film deposition to create the electrical contacts for electrospray.
Integration of all components on the chip provides well-defined peaks that are two to three times narrower than those produced by conventional nanocolumn systems. Small elution volume and enhanced peak height provide greater sensitivity. Zero-dead-volume connections deliver the full separation efficiency of the analytical column to provide better resolution of more compounds. Peak dispersion is virtually eliminated, resulting in narrower, better-defined peaks and improved separations. This chip-based approach improves overall robustness, reliability, and ease-of-use.
A specialized interface contains the loading mechanism for chip positioning and the microvalve for nano-LC connections and flow switching, and it connects to the mass spectrometer. This interface automatically and optimally positions the chip orthogonal to the MS inlet, and it also makes the necessary electrical and fluid connections to the chip. The interface process is automated, requiring users only to slide the chip into the port. The interface module is also fully integrated and controlled by an existing HPLC system.
This chip and interface design provides greatly improved sensitivity, robustness, reliability, and ease-of-use. A range of column chemistries and mass spectrometers have been matched with the chip for applications such as protein identification and quantitation, analysis of PTMs, intact protein analyses, and the analysis of pharmaceuticals.
Protein Identification
LC combined with tandem mass spectrometry (MS–MS) using ion trap or Q-TOF is a key technique for identifying proteins in complex biological samples. Proteins are digested into peptides, which are separated and subjected to MS–MS analysis, and the proteins are then identified from the fragmentation spectra by searching comprehensive protein databases.

Figure 2
Often, proteins of interest are present at very low abundance, so the analytical methods must be extremely sensitive. Nanoflow LC is used most often for protein identification, due to its superior sensitivity over conventional HPLC. In a comparison of a conventional nanoflow system and HPLC chip-MS, analyzing a protein spot extracted from an SDS gel, the number of identified peptides and the resultant number of identified proteins were significantly higher with this system, even when tested against a longer nanocolumn than that used on the chip (Figure 2). This superior performance is due to elimination of dead volumes and minimal sample adsorption on the polyimide chip, as well as integration of the electrospray emitter on the chip, which reduces postcolumn peak dispersion.
Protein Quantitation
While LC–MS is now a well-established technique for biomarker discovery, it also can be used for the confirmation of candidate biomarkers. Multiple reaction monitoring (MRM) on a triple-quadrupole mass spectrometer provides superior sensitivity and selectivity for this application, particularly when paired with this system. In an example with a test peptide, an HPLC-chip–QQQ system easily detected a luciferase peptide at 45 amol, with excellent precision. Using an external standard, a linear calibration curve with a dynamic range of more than four orders of magnitude can be constructed to provide accurate and reliable quantitation (Figure 3). Successful and sensitive biomarker confirmation requires selection of the best ions to use for quantitation, and software can be used to perform theoretical digests of proteins and generate lists of peptides most suitable for sensitive MS analysis, as well as appropriate precursor and product ions for MRM experiments. While biomarker confirmation often is performed with immunoassays, this HPLC chip and MRM approach is a reliable alternative, without the extensive development time often necessary for immunoassays.

Figure 3
Intact Protein Analysis
The routine analysis of biopharmaceutical proteins, as well as purified proteins used in basic research, can be performed reliably with very small amounts of sample using the system. For example, recombinant monoclonal antibodies (mAbs), which have therapeutic applications, are relatively stable biomolecules, but a number of chemical modifications and degradation reactions can occur during their manufacture, formulation, and storage. Accurate mass measurement of the intact antibodies and their subunits can be useful for rapid verification of their integrity.

Figure 4
The chip-based system was coupled to a Q-TOF mass spectrometer to analyze low nanogram levels of intact mAb. Deconvolution of the mass spectrum revealed three main forms of the mAb complex (Figure 4). The peak with a mass of 148,812.81 Da corresponded to the mAb with two units of glycan (G0F) attached, and was determined with a mass accuracy of 5.7 ppm. The peak with a mass of 147,367.94 represents the mAb with only one unit of glycan attached, and the 145,922.00 Da peak is the mAb with no glycan attached. Thus, the high sensitivity and sub-10 ppm mass accuracy of the HPLC-chip–Q-TOF MS system can enable rapid identification of protein heterogeneity and confirmation of glycosylation states, while requiring almost 1000 times less sample than conventional capillary HPLC methods.
Protein Glycosylation
Glycoproteins are involved in immune defense, cell growth, and cell–cell adhesion, and the glycans that help mediate these functions can take on myriad complex structures, requiring analysis techniques that can elucidate them. The chip-based LC–MS system system can be used to identify potential glycoprotein biomarkers in complex protein mixtures, as well as to characterize glycoprotein drugs such as antibodies during quality control. An HPLC chip containing a porous graphitized column was designed specifically for glycan and oligosaccharide separations.
For biomarker analysis, the glycan residues were cleaved enzymatically from human serum proteins, then separated into three fractions (neutral, neutral plus acidic, and acidic) by solid-phase extraction. The resulting fractions were then analyzed on the HPLC chip interfaced with a TOF–MS system. More than 200 possible N-linked oligosaccharides were determined for each of the three fractions (3). Triplicate digestions and multiple injections to the MS system demonstrated excellent reproducibility. A higher capacity chip (four times higher trapping capacity) also gave highly reproducible results. The chip-based LC–MS system shows great promise as a tool for glycan and glycoprotein biomarker discovery.
More than 90% of the protein drugs in existence are glycoproteins. For instance, many different recombinant forms of immunoglobulins (IgGs) are produced as therapeutic glycoprotein drugs for treating life-threatening conditions such as metastatic breast cancer and non-Hodgkin's lymphoma. The efficacy of a glycoprotein drug depends upon the type and extent of glycosylation of the protein. In turn, the degree and types of glycosylation are strong functions of the expression system and the cell culture conditions used in the production of the antibody. Thus, it is critical to characterize the glycan portion of recombinant IgGs as an essential part of antibody quality control.
This HPLC-chip system was coupled with an ion-trap mass spectrometer to characterize N-linked glycans released from IgG (4). Using an on-column amount equivalent to glycans from 1 pmol of starting glycoprotein, this system can be used to detect and identify minor IgG variants. The known major glycans in IgG, designated G0, G1 and G2, were detected, along with versions of all three containing an additional sialic acid residue or bisected GlcNAc, and a fucosylated bisected version of G2 (Figure 5). This system can serve as a platform for IgG quality control assays.

Figure 5
Protein Phosphorylation
Reversible protein phosphorylation plays an important role in cell signaling pathways as well as regulating many other diverse cellular processes. For example, the extracellular signal-regulated kinase (ERK) pathway is essential to the transmission of signals from many extracellular agents to regulate proliferation, differentiation, and cell cycle progression. Signaling via the ERK cascade is mediated by sequential phosphorylation and activation of protein kinases in the different tiers of the cascade. The two key regulatory phosphorylation sites in the human ERK1 protein have been identified as the neighboring T202 and Y204. The degree of phosphorylation of these two sites plays a key role in the overall function of ERK1 in the signal transduction pathway.
However, phosphoprotein analysis to assess phosphorylation levels can be challenging due to small sample sizes, as well the low stoichiometry of phosphorylation. A sensitive MRM assay based upon an HPLC-chip system and utilizing a C18 reversed-phase column coupled to a triple-quadrupole mass spectrometer was developed to quantify the percentage of phosphorylation of T202 and Y204, by reference to 13C-labeled peptides (5). The four peptides, each containing both phosphorylation sites, are without phosphorylation (TY), with single phosphorylation at T202, with single phosphorylation at Y204, and with double phosphorylation at both T202 and Y204. Appropriate precursor and product ion transitions were selected, and calibration curves were developed to allow determination of the molar ratio of each phosphorylation form, with relative standard deviations (RSDs) less than 15% (Table I).
Selective enrichment of phosphorylated proteins and peptides is preferred for LC–MS analysis, and one of the most commonly used methods for enrichment is titanium dioxide (TiO2) adsorption. A new HPLC chip has been developed with titanium dioxide particles flanked on both sides with C18 reversed-phase material (6). This configuration provides researchers with three modes of peptide analysis: standard peptide analysis, phosphopeptide analysis only, and combined peptide and phosphopeptide analysis.
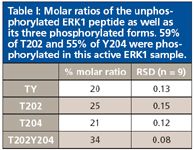
Table 1
Drug Metabolism and Pharmacokinetics (DMPK)
The pressure to reduce costs by saving active pharmaceutical ingredients is driving the industry to scale down laboratory processes, particularly in the DMPK laboratory. During PK studies, the potential lead compound is administered to animals and blood is taken over time to monitor drug adsorption and decay using quantitative LC–MS. The study cost is reduced by using smaller animals, or the recently proposed "single animal" testing, but this significantly limits the sample size for analysis, because one animal must be bled several times. This is an ideal application for the chip-based LC–MS system discussed here.
An ultrahigh capacity (UHC) chip has been developed for this purpose, with a 500-nL enrichment column that provides for concentration of analytes. In addition to improving sensitivity 100 times compared with conventional LC–MS, putting this sample concentration step on the chip increases throughput by reducing the off-chip sample preparation steps to just protein precipitation and centrifugation. This UHC chip system allows quantification of pharmaceuticals in the small volumes of blood required by DMPK.
The challenge presented by analysis of pharmaceuticals is the wide range of polarities. No loss of molecules can be afforded during the on-chip enrichment process. Analysis of a mixture of four pharmaceutical compound (atenolol, atropine, metropolol, and imipranine) covering a wide polarity range (logP 0–4) was achieved using the UHC HPLC-chip system coupled to a triple-quadrupole mass spectrometer (7). A maximum RSD of 0.2% was obtained for the most hydrophilic compound, atenolol, while the RSDs for the rest were under 0.1%. Chip-to-chip reproducibility, using eight runs per chip across 11 chips, gave RSDs below 1%. Most importantly, plasma samples containing only 10 fg/µL of each compound gave prominent peaks that were well above the lowest limit of detection (Figure 6).

Figure 6
Conclusion
Various HPLC-chip chemistries have been integrated with a range of mass spectrometers to provide sensitivity, selectivity, mass accuracy, and precision for a number of applications (8,9). The benefits of chip-based LC–MS are certain to drive the development of more new applications in the near future.
Christine Miller is a Senior Applications Scientist, and Dayin Lin is a Product Manager at Agilent Technologies, Santa Clara, California.
References
(1) H. Yin, et al., Anal. Chem. 77, 527–533 (2005).
(2) M.H. Fortier, et al., Anal. Chem. 77, 1631–1640 (2005).
(3) C.S. Chu, et al., Global NanoLC microchip profiling of human serum oligosaccharides, Oral Presentation at the 2007 Meeting of the American Society for Mass Spectrometry.
(4) P.D. Perkins, Glycoprotein Characterization, Agilent Application Note 5989-5157EN (2006).
(5) N. Tang and C.A. Miller, Quantitation of Protein Phosphorylation Using Multiple Reaction Monitoring, Poster Presentation at the 2008 Meeting of the American Society for Mass Spectrometry.
(6) S. Mohammed, et al., J. Proteome Res.7, 1565–1571 (2008).
(7) S. Buckenmaier, et al., The Column, 20–25 (2008) www.thecolumn.eu.com.
(8) G.O. Staples, et al., Proteomics 9, 1–10 (2009).
(9) A.M. Giessing, et al., RNA 15, 327–336 (2009).

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
The Fundamental Role of Advanced Hyphenated Techniques in Lithium-Ion Battery Research
December 4th 2024Spectroscopy spoke with Uwe Karst, a full professor at the University of Münster in the Institute of Inorganic and Analytical Chemistry, to discuss his research on hyphenated analytical techniques in battery research.
Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.