Savitzky–Golay Smoothing and Differentiation Filter of Even Length: A Gram Polynomial Approach
In various fields such as signal processing, imaging processing, analytical chemistry, and spectroscopic analysis, smoothing and differentiation is important and necessary. With a matrix approach, the Savitzky–Golay smoothing and differentiation filter was extended recently to even length. In this article, a more general approach is proposed for convenient computation.
In the classic article written by Savitzky and Golay (1), which has been cited more than 3800 times according to ISI Web of Science, a kind of digital filter for smoothing and differentiation was developed. In their approach, each successive subset of 2m + 1 points is fitted by a polynomial of degree n (n ≤2m) in the least-squares sense. The s-th (0 ≤ s ≤ n) differentiation (zeroth differentiation = smoothing) of the original data at the midpoint is obtained by performing the differentiation on the fitted polynomial rather than on the original data. Finally, the running least-squares polynomial fitting can be performed simply and automatically by convolving the entire input data with a digital filter of length 2m + 1.
The history and development of the Savitzky–Golay (SG) smoothing and differentiation filter have been reviewed in brief (2). The original tabular coefficients of the SG filter have been corrected by Steinier et al. (3) and Madden (4) subsequently. The typo of a formula by Madden (4) has been spotted recently (5). As pointed out by Mark and Workman (5), one limitation is that the existing SG filter is subject to using an odd number of data subset. With a matrix approach, the SG filter was extended recently for even number (2m) data (i.e., the SG filter could have an even length [6]). The feasibility of the even-length SG filter has been validated with computer simulations (6). In the least-square strain estimator (LSQSE) of ultrasound elastography, it was concluded that a data subset of number 8 could obtain the best performance (7). This method is equivalent to the SG differentiation filter with a linear fitting (8), and the data subset of even number corresponds to an even length of the filter. As discussed in (6), for an even order differentiation, the SG filter of even length has a better characteristic of attenuation at the high frequency of ω= π than that of odd length. However, the matrix approach (6) is not very convenient to calculate the filter coefficients. In this article, a more general approach based upon Gram polynomials is developed to calculate the convolution coefficients of the even-length SG filter for all polynomial degrees, all differentiation orders, and all data points.
Matrix Approach
Consider an even number (2m, 2m – 1 ≥n) of data subset and assume the index of the data samples ranges from –m + 1 to m. The data samples can be regarded equivalently as being symmetrical about the origin, by shifting the origin one half. Therefore, the input signal are represented by (6)
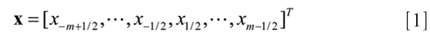
The matrix G(2m) x (n+1) contains the convolution coefficients of the SG filter for different order differentiation at the origin (that is, the imaginary midpoint or the center of symmetry), given by (6)
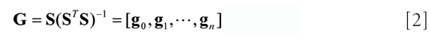
where n is the fitting polynomial degree, S is the 2m x n + 1 basic matrix, as (6)
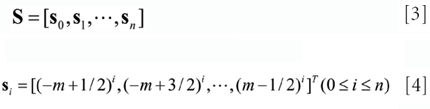
where si is the i-th column vector of S.
The convolution coefficients for the s-th differentiation evaluated at the imaginary midpoint are given by (6)
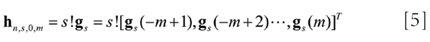
where gs is the s-th column vector of G, the subscript 0 represents the evaluated midpoint. With the matrix approach mentioned earlier, some closed-form solutions as well as the numerical table can be obtained (6). However, only the filter coefficients for the imaginary midpoint are convenient to obtain.
Gram Polynomial Approach
It is expected that it is difficult to obtain the matrix inversion (ST S)-1 in Equation 2 with an explicit formula if s is large. Therefore, one would prefer numerical computation to symbolic computation. If n or s increase, however, the matrix ST S tends to be ill conditioned (9). Hence, a more successful and general approach with the orthogonal polynomials (Gram polynomials or Legendre polynomials) has been proposed (9–11). In this article, such an approach also is extended for an even-length SG filter. All the equations are similar to those in (10), but the effects of the even number data and the aforementioned translation of the origin have been taken into account.
The smoothing and the differentiation are expressed as
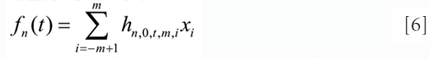
and

respectively, where fn (t) and fn (s) (t) are the smoothing value and the s-th (1 ≤s ≤n) differentiation value evaluated at position t, with polynomial order n and data number 2m; xi is the original data value at point i before shifting the origin (–m + 1 ≤ i ≤m); and hn,0,t,m,i and hn,d,t,m,i are the corresponding coefficients for smoothing and differentiation, respectively. Note that t = 0 for the imaginary midpoint of the data subset, while t = –m + 1/2, –m + 3/2 and m – 1/2 for the first, second, and last points, repetitively.
The convolution coefficients in Equations 6 and 7 are calculated as
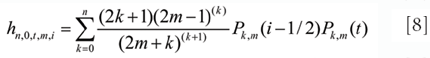
and

respectively, where (a)(b) is a generalized factorial function given by
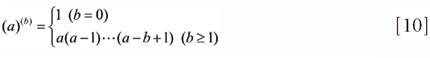
Pk,m (t) is the Gram polynomial of order k, over 2m points (symmetrical about the origin), evaluated at position t. It is defined by
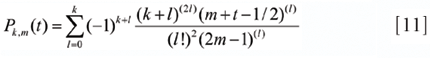
P(s)k,m is the s-th derivate of the Gram polynomial evaluated at position t, that is,

According to the ordinary definition of the Gram polynomials and their derivatives given by Equations 11 and 12, respectively, the explicit expressions for some low-order Gram polynomials are calculated and listed as follows
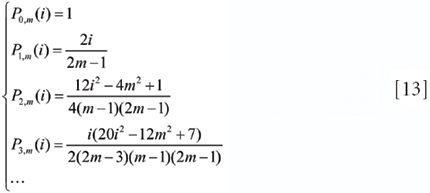
However, Equations 11 and 12 are not very practical for calculation purposes, especially for calculating the derivatives. An alternative method using the recursive properties of the Gram polynomials and their derivatives can be expressed as

and
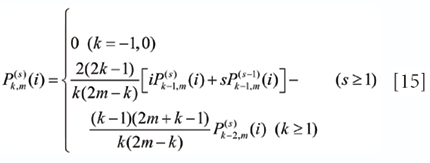
respectively.
Results and Discussion
The appendix contains the routines of the previously mentioned approach with Gram polynomials. With these routines, one easily can obtain results that are identical to those given by (6). Table I lists some results of the even-length SG filter for the midpoint of the data subset. Furthermore, the results in (6) or Equation 5 are only for the midpoint. Using Equations 8 and 9, the even-length SG filter evaluated at any position can be obtained easily. Note that t = 0 for the imaginary midpoint of the data subset, while t = –m + 1/2, –m + 3/2 and m – 1/2 for the first, second, and last points, repetitively. Some convolution coefficients of the even-length SG filters evaluated at the sampled data points as well as those at the imaginary midpoint are given in Table II.

Table I. Convolution coefficients of the even-length SG filter (t = 0)
As can be seen in Table II, the SG filters for the midpoint are symmetrical (or antisymmetical), and therefore, have the advantage of linear phase. In addition, the asymmetric odd-length SG filters (for points except the midpoint) can distort data and have poorer noise attenuation (12), especially for the points far away from the the midpoint. They are useful in some cases, but should be applied with caution (12). It is expected that the asymmetric even-length SG filters will have some similar performances.

Table II. Convolution coefficients of the even-length SG filter (m = 3)
The even-length SG filter for the midpoint is similar to the interpolation in the sense of least square polynomial fitting (2m – 1 > p) or polynomial approximation (2m – 1 = p). Therefore, the results of smoothing and differentiation correspond to an imaginary point between the original sampled data points, rather than the same wavelength as one of the original data. To avoid the asymmetry, the even-length SG filter evaluated at the midpoint also can be used for the neighboring sampled points. However, that will result in a time delay of half a sampling interval and a worse performance accordingly (6). For example, the two-point backward derivative formula, which can be regarded as an even-length SG differentiation filter (6), is more precise to calculate the derivative at a point halfway between the data points, rather than at either one of the two points. Fortunately, it does not matter when the sampling interval is small enough.
Conclusion
The widely used SG filter was extended recently for even number data subset. In this article, a more general approach with Gram polynomials is proposed to calculate the convolution coefficients of the even-length SG filter for all polynomial degrees, all differentiation orders, and all evaluated points.
Appendix
The routines of the even-length SG filter written in MATLAB (The MathWorks, Inc., Natick, MA) can be found in (13). The routines of the Gram polynomial approach presented in this paper are given as below. They also are written in MATLAB and can obtain both of the numerical results and the symbolic results (by using the Symbolic Math Toolbox). However, the symbolic calculation is much slower.
function hnstm = gsdf_even_gram_poly (n,s,t,m)
% hnstm = sgsdf_gram_poly(n,s,t,m)
% n: polynomial degree
% s: derivative(differentiation) order (0 = smoothing)
% t: evaluation point (commonly, t = 0, i.e., smoothing or differentiation
% at the imaginary central point of the 2*m points)
% 2*m: data point number, i.e., filter length
% hnstm: convolution coefficients
hnstm = [];
for i = -m + 1/2:m - 1/2
hnstm = [hnstm hinstm(i,n,s,t,m)];
end
function value=hinstm(i,n,s,t,m)
% Equs. (8) and (9)
value=0;
for k=0:n
value=value+(2*k+1)*gff(2*m1,k)/gff(2*m+k,k+1)*...
Pkmi(k,m,i)*Pkmsi(k,m,s,t);
end
function value = gff(a,b)
% generalized factorial function (Equation 10)
if b == 0
value = 1;
else
value = prod(a-(0:b - 1));
end
function value = Pkmsi(k,m,s,i)
% s-th derivative of Gram polynomials (Equation 15)
if s == 0
value = Pkmi(k,m,i);
else if k == -1 || k == 0
value = 0;
else
value = 2*(2*k-1)/k/(2*m-k)*(i*Pkmsi(k-1,m,s,i)+s*Pkmsi(k-1,m,s-1,i))-...
(k-1)*(2*m+k-1)/k/(2*m-k)*Pkmsi(k-2,m,s,i);
end
function value = Pkmi(k,m,i)
% Gram polynomials (Equation 14)
if k == -1
value = 0;
else if k == 0
value=1;
else
value = 2*(2*k-1)/k/(2*m-k)*i*Pkmi(k-1,m,i)-(k-1)*(2*m+k-1)/k/(2*m-k)*Pkmi(k-2,m,i);
end
The closed-form solutions listed in (2) can be obtained with the following routine.
function sgsdf_even_closed_form(n,s)
% n: polynomial degree
% s: derivative(differentiation) order (0 = smoothing)
subs(factor(hinstm(sym('k'),n,s, sym(0), sym('m'))),'k', 'k-1/2')
Some examples of the usage of these routines are given in Table III.

Table III. Some examples of the usage of the MATLAB routines
Acknowledgment
This work is supported in part by the National Natural Science Foundation of China (60171039, 30470466).
References
1. A. Savitzky and M.J.E. Golay, Anal. Chem. 36(8), 1627–1639 (1964).
2. J.W. Luo, K. Ying, P. He, and J. Bai, Digit. Signal Prog. 15(2), 122–136 (2005).
3. J. Steinier, Y. Termonia, and J. Deltour, Anal. Chem. 44(11), 1906–1909 (1972).
4. H.H. Madden, Anal. Chem. 50(9), 1383–1386 (1978).
5. H. Mark and J. Workman, Spectroscopy 18(12), 106–111 (2003).
6. J.W. Luo, K. Ying, and J. Bai, Signal Process., 85(7) 1429–1434 (2005).
7. F. Kallel and J. Ophir, Ultrason. Imaging 19(3), 195–208 (1997).
8. J.W. Luo, J. Bai, P. He, and K. Ying, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 51(9), 1119–1127 (2004).
9. J.S. Lim, and A.V. Oppenheim, Eds., Advanced Topics in Signal Processing (Prentice-Hall, Englewood Cliffs, NJ, 1988).
10. P.A. Gorry, Anal. Chem. 62(6), 570–573 (1990).
11. J.S. Waugh, Ed., Advances in Magnetic Resonance (Academic Press, New York, NY, 1966), vol. 2.
12. P.D. Wentzell, T.P. Doherty, and S.R. Crouch, Anal. Chem.s 59(2), 367–371 (1987).
13. The MATLAB Central File Exchange, at: http://www.mathworks.com/matlabcentral/fileexchange/loadCategory.do?objectId=66&objectType=Category.
Jianwen Luo and Jing Bai are with the Department of Biomedical Engineering at Tsinghua University (Beijing, China). E-mail: jianwen.luo@gmail.com

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
The Fundamental Role of Advanced Hyphenated Techniques in Lithium-Ion Battery Research
December 4th 2024Spectroscopy spoke with Uwe Karst, a full professor at the University of Münster in the Institute of Inorganic and Analytical Chemistry, to discuss his research on hyphenated analytical techniques in battery research.
Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.