Using High-Resolution LC–MS to Analyze Complex Sample
Special Issues
The authors discuss the use of high-resolution LC-MS to analyze complex samples in regulated environments such as food and animal-feed analysis.
Identifying and quantifying analytes in highly complex samples, such as extracts from food, animal feed, pesticides, mycotoxins, and veterinary drugs, is a major analytical challenge for mass spectrometry. Accurate screening of these samples is critical in regulated environments such as food and animal-feed analysis.
Triple-quadrupole mass spectrometers have been used for identifying and quantifying analytes in complex samples, though that approach comes with a number of limitations, including the inability to re-interrogate the data postacquisition, as well as the inability to interrogate unknowns and the limited number of compounds per analysis. Time-of-flight (TOF) instruments have also been used for this application, but the technology's low resolution (15,000) has led to inaccurate mass measurements caused by unresolved background matrix interferences.
A growing record of successful experiments has shown that the ability to perform high-resolution liquid chromatography–tandem mass spectrometry (LC–MSn) analyses is the key to measuring accurate mass, determining elemental compositions, and performing highly specific quantification on highly complex samples.
While the advantages of high resolution have been demonstrated, there remains significant confusion in the scientific community about what constitutes a high-resolution approach to MS. This article will attempt to clarify the terms, technology, and methods used in high-resolution MS and then present a brief case study that shows the application of a high-resolution LC–MSn approach in greater depth.
Defining "High Resolution"
The application of high-resolution technology in MS is fundamentally the same as in photography: a high-resolution photograph shows great detail; a low-resolution photograph is blurry. However, the terms high (mass) resolution and accurate mass are often (incorrectly) used interchangeably. High resolution is necessary to separate peaks from one another and to ensure that ions of only one kind contribute to a particular measurement. The measurement may be an accurate mass determination or a highly specific quantification.
High resolution is particularly important for experiments involving complex mixtures, such as samples generated from a matrix (for example, biological, environmental), because these will often contain a significant number of background ions. In such cases, high resolving power will make the difference between detecting analytes at low concentration or not detecting these analytes due to the masking effect of isobaric matrix interferences.
Because two terms, resolution and resolving power, are used to characterize mass spectrometers, it is useful to look at their definitions.
The International Union of Pure and Applied Chemistry (IUPAC) defines resolving power in MS as
"The ability to distinguish between ions differing in the quotient mass/charge by a small increment. It may be characterized by giving the peak width — measured in mass units, expressed as a function of mass — for at least two points on the peak, specifically at 50% and at 5% of the maximum peak height."
The IUPAC defines resolution in MS (m/Δm) as
"10% valley definition: Let two peaks of equal height in a mass spectrum at masses m and m – Δm be separated by a valley, which at its lowest point is just 10% of the height of either peak. For similar peaks at a mass exceeding m, let the height of the valley at its lowest point be more (by any amount) than 10% of either peak height."
"Peak-width definition: For a single peak made up of singly charged ions at mass m in a mass spectrum, the resolution may be expressed as m/Δm, where Δm is the width of the peak at a height that is a specified fraction of the maximum peak height." It is recommended that one of three values — 50%, 5% or 0.5% — always be used. For an isolated symmetrical peak recorded with a system that is linear in the range between 5% and 10% of the peak, the 5% peak-width definition is technically equivalent to the 10% valley definition. A common standard is the definition of resolution based upon Δm being full width of the peak at half its maximum height, sometimes abbreviated FWHM. Preferably, this abbreviation should be defined the first time it is used.
The terms resolution and resolving power are basically interchangeable. In fact, both resolution and resolving power are defined as R = (m/Δm). The difference, however, is how Δm is defined.
Resolution of magnetic-sector instruments is normally given according to the 10% valley definition, which defines Δm as the mass difference between two resolved peaks, with a 10% valley between them.
All other definitions for resolution are measured on a single peak. Because there are several ways to do this, one should always look at the definition actually used before comparing numerical values. For example, the 10% valley definition, where Δm is based on a real peak separation, is equivalent to a single peak measurement, where Δm is defined by the peak width at 5% peak height, as can be seen below.
The most commonly used method to measure the resolution of quadrupole MS, Fourier transform ion cyclotron resonance (FTICR), Orbitrap, and TOF MS follows the FWHM definition, which uses the width of a peak at 50% of its height as a measure for Δm.
The numerical value arrived at using the FWHM definition is naturally always larger than the value one would arrive at using the "width at 5% peak height" (10% valley) definition. In fact, the numerical FWHM resolution necessary to achieve mass separation to 10% valley is about twice the value, according to the 10% valley definition. This means that a resolution of 20,000 (FWHM) is equivalent to a resolution of 10,000 (10% valley).
An instrument that has a resolution of 10,000 (FWHM) will separate ions at m/z 500.0 from ions at m/z 500.1 (and not 1000.0 from 1000.1).
In the above example, where resolution values are given according to the FWHM definition, the baseline separation of CO from N2 is shown (right-hand panel), which would require a resolution of 2300 following the 10% valley definition. However, at an instrumental resolution of 2300 (FWHM), the individual CO and N2 peaks (green and blue) merge to give the signal outlined by the thick black line. The instrumental resolution must be increased to about 5000 (FWHM) to separate and accurately measure the mass of each peak.
Instrument Types and Mass Resolution
In all following examples, "resolution" and "resolving power" will be used synonymously and numerical values will follow the FWHM definition.
In the case of quadrupole ion trap instruments (such as the Thermo Scientific LTQ mass spectrometers) and transmission quadrupole instruments (such as the Thermo Scientific TSQ Quantum mass spectrometers), the instrumental resolution increases linearly with mass — that is, if the instrument is set to separate one integer from its neighbor (for example 100 from 101), then the resolution will be increased from 200 at m/z 100 to 2000 at m/z 1000. This resolution setting is called "unit mass resolution."
TOF and double-focusing mass spectrometers (for example, Thermo Scientific DFS) operate at constant resolving power. This means that the set resolution will be the same for the entire mass range.
Thermo Fisher Scientific FTICR and Orbitrap series mass spectrometers operate at varying resolving power. In the case of FTICR, the resolution decreases linearly with mass; for Orbitraps, the resolution decreases with the square root of mass. For example:
Orbitrap MS: R = 100,000 at m/z 400 becomes R = 50,000 at m/z 1600
FTICR MS: R = 100,000 at m/z 400 becomes R = 25,000 at m/z 1600
The Importance of High Resolution: A Case Study
High-resolution MS is generally used either to measure accurate masses (and thereby enable the determination of elemental compositions) or to enable highly specific quantification. In either case the mass centroid of a peak profile is measured. For a quantification experiment, unambiguous results can be obtained only if sufficient resolving power is used to separate the target analyte from any possible interference. If the resolving power of the instrument is insufficient, it will result in false-positive or false-negative signal responses.
To demonstrate the utility of high-resolution MS and clarify how this analysis works in practice, a horse-feed extract spiked with a mixture of 116 pesticides was analyzed as an example of an extremely complex matrix. The work demonstrates a full-scan screening approach utilizing resolutions of up to 100,000. The experiment reveals the advantages of high resolution in resolving coeluted, isobaric target compounds and determining their elemental composition.
A dilution series ranging from 2 to 250 ppb (for each compound) was measured in duplicates at two different resolution settings. In addition a 100-ppb sample of the same mixture was analyzed at a resolution of 50,000 to determine the maximum number of detectable substances for this method.
Methods
A non-hybrid single stage Orbitrap mass spectrometer (Exactive, Thermo Fisher Scientific, Bremen, Germany) coupled to a ultrahigh-pressure liquid chromatography (UHPLC) system (Thermo Scientific Accela, Thermo Fisher Scientific, San Jose, California) was used to evaluate a highly complex mixture of 116 pesticides, mycotoxins, and plant toxins in different concentrations. A 12-min gradient was applied to a 50 mm × 2 mm RP C18 column (Thermo Scientific Hypersil GOLD with 1.9-μm particles, Thermo Fisher Scientific, Madison, Wisconsin) with water–acetonitrile eluents.
The method developed was evaluated with respect to sensitivity, selectivity, and linearity in standard solutions and extracts from animal feed. Mass measurements were performed at different resolution settings (R = 15,000 and R = 50,000) to enable comparisons to data acquired by TOF instruments and to demonstrate the advantage of ultrahigh resolution. Orbitrap detection was carried out using automatic control of the number of ions entering the detector (AGC, target value = 106).
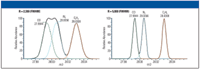
Figure 1: Separation of isobaric peaks at different resolution settings.
Results
Resolution of Isobaric Pesticides
In cases where isobaric compounds are coeluted, erroneous mass accuracy and elemental composition assignment will occur if the resolving power of the mass spectrometer is insufficient to separate these compounds. Figure 2 shows two pesticides, thiamethoxam (C8H10CIN5O3S) and parathion (C10H14NO5PS), which have protonated molecular ions (MH+) at 292.02656 and 292.04031, respectively. A resolution higher than 40,000 is needed to resolve the protonated molecular ion of these two compounds completely. This is a prerequisite for analysis of low concentration compounds in the presence of higher abundant ones. The example in Figure 2 shows an approximate 1:3 mixture of both pesticides measured and simulated.
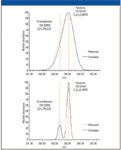
Figure 2: Mass chromatogram of two isobaric pesticides measured at a resolution of 20,000 (top) and 50,000 (bottom). Superimposed is the simulated mass trace at each resolution setting.
Influence on Elemental Composition Determination
A limited resolution of 15,000 results in two major limitations. First, the detection of unresolved doublets may result in significant mass errors that are outside the characteristic accuracy specification of the Exactive instrument. As a consequence, at lower resolution settings the mass windows for elemental composition determination have to be increased, resulting in much larger numbers of elemental composition proposals for the unknown or targeted compounds.
This can be seen in the example (Figure 3) of pirimicarb at m/z 239.1503. Due to the presence of an isobaric interference, the peak at 239 shows a mass error of 6.5 ppm. At a resolution of 15,000, the underlying interference causes an apparent shift to higher mass, whereas at higher resolution (here 80,000), the doublet is clearly resolved and the mass accuracy is well within instrument specifications.
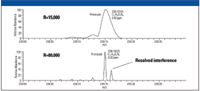
Figure 3: Improved mass accuracy simply by increasing the resolution in order to resolve the doublet. The peak at 15,000 resolution splits up into two by increasing the resolution to 80,000.
To limit the number of candidate elemental compositions to a single confident assignment, a sophisticated software algorithm is used. It takes into account the peak height and mass accuracy of the monoisotopic peak and its isotopes. However, to function correctly, all of the accurate mass values for the isotopic peaks must be within specified limits. The absence of interference peaks can only be ensured by use of high resolution (as can be seen in Figure 4). Here the fungicide azoxystrobin is shown at resolutions of 15,000 and 80,000. The medium resolution spectrum shows very good mass accuracy for the monoisotopic peak but gives unusually high mass errors for the A+1 and A+2 ions.
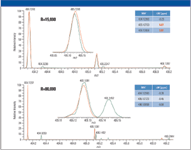
Figure 4: The importance of resolution for the molecular ion and its isotopes. At high resolution, the complete molecular ion cluster is detected correctly.
This is due to an interference at m/z 405.1452, which cannot be resolved at medium resolution. The high resolution spectrum shows excellent mass accuracy for all three measured isotopes. Determining elemental compositions using data acquired at ~15,000 resolution will result in misleading or incorrect data. Only sufficient high resolution allows the determination of the accurate mass of the complete molecular ion cluster and therefore allows automated assignment of an elemental formula with a high degree of confidence.
LC–MS analysis of the extracted spiked samples showed the presence of 95 out of 116 compounds at 100 ppb in matrix. Figure 5 shows the overlaid ion chromatograms for all 116 compounds (3 ppm window) at 50 ppb (in matrix). The number of recovered pesticides in different concentrations is shown in Figure 5. The data illustrate a greater number of detected compounds (higher sensitivity) with an extraction window of 3 ppm at a higher resolution setting. This is exemplified in Figure 6, where extracted ion chromatograms of sulcotrion at 50 ppb for R = 15,000 and R = 50,000 are shown. The higher resolved spectrum displays two peaks, of which the smaller one is sulcotrion.
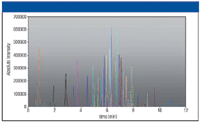
Figure 5: Overlaid extracted ion chromatograms from a mixture of 116 pesticides and mycotoxins at a 100 ppb level. Extraction was done with 3 ppm mass window. The inset chart shows the number of detected compounds at different concentrations (in matrix) at two different resolution settings.
The lower resolved peak masks the pesticide signal completely. The only indication for the presence of sulcotrion is a slightly broader peak or shoulder and a mass shift of the interfering ion toward higher masses. This would lead to a false-negative result if the analysis was only performed at a resolution of 15,000, and is the major reason the number of identified components in the case of R = 15,000 decreases disproportionately to the measurements at higher resolving power (diagram, Figure 6).
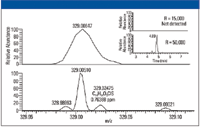
Figure 6: Expanded view of the pesticide mixture at different resolution settings (top: 15,000; bottom: 50,000). Pesticide Sulcotrion (m/z 328.02475) is masked under background ions at a resolution of 15,000 but is easily detected at 50,000 resolution (see also mass chromatogram inset).
For this reason, a dilution series was measured at higher resolution settings: 2 ppb, 10 ppb, 50 ppb, and 250 ppb. The results showed excellent linearity down to the 2 ppb, creating peaks that were essentially indistinguishable from each other.
Conclusions
High resolution LC-MSn analyses (with resolving power up to 100,000 using Orbitrap technology) provide precise mass accuracy for complex sample analysis and provide excellent sensitivity, linearity, and selectivity in multi-residue screening of complex matrices, as instruments with this level of high resolution demonstrate. The low resolution inherent in TOF technology introduces significant risk of inaccurate mass measurements caused by an unresolved background matrix, a critical concern in regulated environments where a false negative can have serious health consequences.
Markus Kellman, Andreas Wieghaus, and Helmut Muenster are with Thermo Fisher Scientific, Bremen, Germany. Lester Taylor and Dipankar Ghosh are with Thermo Fisher Scientific, San Jose, California.

Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.