Large-Scale Targeted Protein Quantification Using Wide Selected-Ion Monitoring Data-Independent Acquisition
This article describes the development of a new data-independent acquisition (DIA) workflow for protein quantification that uses a mass spectrometer that combines three types of mass analyzers to achieve lower limits of detection (LOD), higher sensitivity, more accurate quantitative results, wider dynamic range, and better reproducibility than existing high-resolution accurate-mass (HRAM) tandem mass spectrometry (MS-MS) DIA workflows.
This article describes the development of a new data-independent acquisition (DIA) workflow for protein quantification that uses a mass spectrometer that combines three types of mass analyzers to achieve lower limits of detection (LOD), higher sensitivity, more accurate quantitative results, wider dynamic range, and better reproducibility than existing high-resolution accurate-mass (HRAM) tandem mass spectrometry (MS-MS) DIA workflows. The combined use of a mass-resolving quadrupole, orbital trap mass analyzer, collision cell, and linear ion trap with appropriate software was shown to minimize interferences caused by product ions from coeluted background compounds and significantly improve the analysis of highly complex samples.
Proteomics studies are rapidly turning from qualitative to largely quantitative experiments to better understand the functions and interactions of proteins in biological systems and verify long lists of putative biomarkers. The extreme complexity and large dynamic range of proteins in standard biological samples challenge traditional data-dependent workflows. They necessitate very fast tandem mass spectrometry (MS-MS) with simultaneous high sensitivity and reproducibility. Without these, it is difficult to achieve the depth of sample interrogation necessary to identify and quantify the same targets in replicate runs to accurately determine differences among samples.
Recently, several data-independent acquisition (DIA) approaches have been explored to increase reproducibility and comprehensiveness for better quantification (1,2). One such approach acquires only high-resolution, accurate-mass (HRAM) MS-MS data (no full scans) generated with very wide isolation windows (~25 m/z). These data are then interrogated for quantification using extracted ion chromatograms of targeted fragment ions only. The quantitative performance from using only HRAM MS-MS data generated with wide isolation windows is potentially compromised by interfering product ions from coeluted background compounds. For highly complex biological samples, a portion of the targeted fragment ions are potentially contaminated by fragment ions from nontargeted peptides that coelute in the same retention time window and are coisolated within a 25 m/z wide window. These interferences can result in higher limits of detection (LOD) and limits of quantitation (LOQ), less accurate quantitative results, and narrower dynamic range (2,3).
Recent MS architecture, which combines a mass resolving quadrupole, an orbital trap mass analyzer, a collision cell, and a linear ion trap mass analyzer, was applied to achieve lower LOD and high selectivity. The orbital trap detector is designed to provide a resolving power of 240,000 at a scan rate of 1.5 Hz. The linear ion trap can collect more than 20 high-quality collision-induced dissociation (CID) MS-MS spectra in 1 s. This architecture enables parallel orbital trap selected-ion monitoring (SIM) scanning with rapid and sensitive targeted ion trap MS-MS detection to generate data with the resolution and sensitivity required for accurate data-independent analyses.
A DIA workflow was developed that takes advantage of this architecture. It simultaneously collects HRAM SIM spectra with wide isolation windows in the orbital trap detector and CID MS-MS spectra in the linear ion trap detector. This method is referred to as wide selected-ion monitoring, data-independent acquisition, or WiSIM-DIA. The WiSIM-DIA method consists of three SIM scans acquired with extremely high resolution and mass accuracy, and with wide isolation windows (240,000 resolution full width at half maximum [FWHM] at m/z 200, and <3 ppm, respectively). It covers all precursor ions between m/z 400 and 1000 while increasing the sensitivity relative to standard full-scan analysis. The use of a wide SIM window for the collection of ions in the first step of this method leads to the selective enrichment of analytes in the selected SIM mass range and enables the detection and quantification of low abundance peptides in the final targeted extraction step. In parallel with each SIM spectrum, 17 sequential ion trap MS-MS spectra with 12 m/z isolation windows are acquired to detect fragment ions across each associated 200 m/z SIM mass range (Figure 1). Quantification of targeted proteins after data acquisition is carried out using extracted ion chromatograms of HRAM SIM data with a ±5 ppm window. Simultaneous peptide sequence confirmations are carried out using CID MS-MS relying on a spectral library.
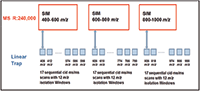
Figure 1: WiSIM-DIA workflow schematic for the collection of HRAM SIM scans at 240k resolution while simultaneously acquiring 17 sequential CID.
The main advantages of the WiSIM-DIA workflow, relative to the wide-window MS-MS-only DIA workflows, are that there is enough time to acquire HRAM precursor data for accurate and sensitive detection and quantification, while the fast and sensitive MS-MS can be used for reliable confirmation. As a result, the quantitative performance of this method does not suffer from the drawbacks of wide-window MS-MS-only DIA workflows. All quantitative analyses are carried out using HRAM SIM data. The CID MS-MS data are used only for peptide confirmation. Therefore, the fragment ion contamination caused from nontargeted, coeluted peptides does not impact the quantitative results. This WiSIM-DIA approach increases selectivity, sensitivity, and reproducibility, ultimately improving the accuracy and throughput of the quantification experiments by using precursor ions collected with very high (240,000) resolving power. The fragment ions detected in the WiSIM-DIA approach, which are used only for confirmation of identity, are from narrower precursor populations (12 m/z), and are detected by a more sensitive mass analyzer. Therefore, this WiSIM-DIA approach can quantify all detected peptide precursor peaks with high sensitivity and selectivity, yielding a complete and reproducible quantitative data set in a single sample injection. In addition, all MS-MS product ion information is recorded for sequence confirmation of any peptide of interest within the mass range of m/z 400–1000 by subsequent confirmatory matching of specific product ions of an easy-to-generate, sample-specific spectral library. The details of this unique WiSIM-DIA workflow and its quantitative performances are reported here.
Experimental
Sample Preparation
Sample 1 (a dilution series for evaluating detection limits and linear dynamic range): A mixture of 14 isotopically labeled synthetic peptides was spiked into a complex E. coli digest (500 ng/μL) at five different concentrations (0.01, 0.1, 1, 10, and 100 fmol/μL) to generate a dilution series.
Sample 2 (for evaluating detection and quantification of low- to high-abundance proteins in a complex mixture): A digest of six proteins with concentrations covering five orders of magnitude (0.01, 0.1, 1, 10, 100, and 1000 fmol/μL) was spiked into an E. coli digest (500 ng/μL).
Sample 3 (a sample set for large-scale relative quantification comparison of separate complex mixtures): An E. coli digest was analyzed at 250 ng and 500 ng on column.
Table I shows the experimental conditions for the nano-LC, MS, and software systems used.

Table I: Experimental conditions
Six experiments were conducted: Three Orbitrap analyzer (Thermo Fisher Scientific) SIM experiments (experiments: 1, 3, and 5) covered the precursor mass range of m/z 400–1000. Each SIM experiment was followed by a targeted MSn experiment, which carried out 17 consecutive CID MS-MS acquisitions using a predefined precursor ion inclusion list (Figure 2). This covered the associated Orbitrap analyzer SIM experiment mass range (experiments: 2, 4, and 6). The instrument method setup can be downloaded at http://planetorbitrap.com.
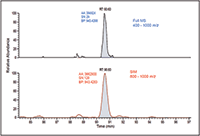
Figure 2: Significant increase in sensitivity using wide-window SIM scans compared to full MS scans, MS-MS scans of 12 m/z isolation.
Data Processing
Dedicated software was used for targeted qualitative and quantitative data extraction after data acquisition. A spectral library containing precursor ion and CID MS-MS information was established using previous discovery data collected on a similar MS system. The extracted-ion chromatograms of isotope 12C and 13C precursor ions per targeted peptide were used for quantification with a ±5 ppm window. The eight most intense fragment ions (b and y types) detected from discovery data for each peptide were used for confirmation through spectral library matching.
Results and Discussion
Decreased Detection Limits with SIM Acquisitions Relative to Full MS Acquisitions
The newly developed WiSIM-DIA workflow provided quantitative results with a label-free approach. Standard, label-free experiments rely on full-scan data for quantification of the precursors; this often compromises the detection of low-intensity ions during the elution of more intense species. The WiSIM-DIA approach was developed to alleviate this issue through the use of sequential SIM acquisitions for quantification. Unlike a full mass-range acquisition, the SIM acquisition with a 200 m/z isolation window effectively "enriches" all ions in that window while excluding all other ions outside the mass range of interest. As a result, the SIM acquisition, even with a 200 m/z isolation window, can provide much higher sensitivity for detecting low-abundance peptides compared to full-mass-range acquisitions. An example of this is shown in Figure 2 where simply using a SIM acquisition allows for a nearly fivefold increase in signal-to-noise ratio. When applicable, the expected retention time per targeted peptide was also considered when extracting the quantitative information.
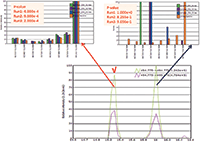
Figure 3: WiSIM-DIA verifies peptide identification using spectral matching. Confident peptide sequence confirmation by extracting the eight most intense fragment ions from the DIA-CID MS-MS data and matching the fragment ion distribution to the CID MS-MS spectrum stored in the spectral library. The peptide eluted at 15.3 min is confirmed to be the correct target with p-values less than 1 Ã 104. The peak with the precursor ion mass at 16 min elution time is a contaminant.
Peptide Sequence Verification Using Ion Trap CID MS-MS
For peptide sequence confirmation, the data-independent CID MS-MS acquisitions with a 12 m/z window over each m/z 200 SIM window were used. Unlike data-dependent analysis that requires triggering of a specific precursor for fragmentation and detection, leading to a lack of reproducibility between replicates, data-independent MS-MS acquisitions are scheduled and cover the complete mass range in every analysis. Eight of the most intense fragment ions (b and y types) identified in discovery (data-dependent) experiments were targeted in the ion trap CID MS-MS data using a ±500 ppm extraction window. They were then used for sequence verification through spectral library matching. A p-value (probability of random spectral matching) was calculated using standard statistical methods to compare the multiple product ion distribution extracted from the DIA MS-MS data with MS-MS data of a peptide stored in a spectra library (4). A smaller p-value represented a better match between the observed data and the spectral library. A peptide with a p-value of less than 0.1 was considered to be identified with high confidence by the library match (3). The ion trap CID MS-MS system provided high sensitivity and good spectral quality without prior knowledge of precursor ion charge state, unlike collision cell fragmentation. It was able to confirm the targeted peptide sequences with sufficient selectivity through the spectral library matches (Figure 3).
Figure 4: WiSIM-DIA workflow provides 10 amol LOD with four orders of magnitude linear dynamic range width in parallel.
Evaluation of Detection Limits and Linear Dynamic Range of the WiSIM-DIA Workflow in a Complex Mixture
The detection limits and the quantitative dynamic range of the unique WiSIM-DIA workflow were evaluated using a dilution series of a mixture of 14 isotopically labeled synthesized peptides spiked into E. coli digests (500 ng/μL) at five different concentrations (0.01, 0.1, 1, 10, and 100 fmol/μL). In addition to the decreased detection limit as a result of using SIM acquisitions, the very high resolving power of 240,000 enabled unambiguous detection of targeted peptide peaks from matrix interferences, even at the lowest 10 amol concentration level. As a result, LODs down to 10 amol were achieved. Table II shows the observed coefficient of variation (CV) of each spiked peptide from triplicate runs at each concentration level. All spiked peptides were detected at the lowest concentration level tested (10 amol). The CVs of all spiked peptides at 100 amol were less than 15% with the exception of one peptide (22%) (Table II). With the low LOD and high selectivity of the DIA workflow, each spiked peptide was able to be detected over four orders of linear dynamic range (Figure 4).
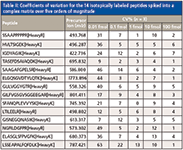
Table II: Coefficients of variation for the 14 isotopically labeled peptides spiked into a complex matrix over five orders of magnitude
Detection and Quantification of Low- to High-Abundance Proteins in a Complex Mixture in a Single Run with the WiSIM-DIA Workflow
To further evaluate the ability of the new WiSIM-DIA workflow to quantify targeted peptides over a wide dynamic range in a single run, six bovine protein digests were spiked into a 500-ng E. coli matrix at an abundance level that spanned over five orders of dynamic range (low-abundance to high-abundance proteins). Among the six spiked proteins, bovine serum albumin (BSA) was spiked at the lowest abundance level (10 amol on column) and beta-lactoglobulin was spiked at the highest abundance level (1 pmol on column) (Table III). The sample was run in triplicate using the WiSIM-DIA workflow. The HRAM SIM acquisitions reproducibly provided low limits of detection, high selectivity, and wide dynamic range for all six spiked proteins over five orders of magnitude of dynamic range. Table III summarizes the detected peptides per spiked protein and observed %CVs. Approximately 90% of quantified peptides gave %CVs less than 10%.

Table III: Detection and quantification of six low- to high-abundance bovine proteins in a complex mixture over five orders of dynamic concentrations in a single experiment
Large-Scale Relative Quantification Using the WiSIM-DIA Workflow
To further evaluate the analytical precision and quantitative accuracy of the WiSIM-DIA workflow when applied to large-scale quantitative experiments, a 500-ng on column E. coli digest and a 250-ng on column E. coli digest were compared. Each sample was run in triplicate using the WiSIM-DIA instrument method for quantification.
For targeted E. coli peptide selection and spectral library generation, a 1-μg E. coli sample was run in triplicate with a standard shotgun data-dependent acquisition (DDA) experiment using the orbital trap analyzer full scan and ion trap CID MS-MS analyses. A database search of the DDA raw files was performed against an E. coli database. The SEQUEST HT search engine was used. A target decoy peptide spectral match (PSM) validator (0.01 FDR strict–0.05 FDR relaxed) was used for PSM validation. The search results of the triplicate runs were combined and 1100 unique E. coli proteins were identified, which included at least two identified peptides with high confidence, number 1 peptide rank, and minimal cross-correlation scores (2.0 for charge 2, 2.25 for charge 3, and 2.5 for charge 4). These peptides were selected as the quantitative targets for targeted extraction in the large-scale quantitative comparison of E. coli digests using the WiSIM DIA workflow.
Search results were used to generate a sample-specific spectral library. Only the MS-MS spectra that passed the minimal cross-correlation scores were imported into the spectral library. All peptides in the spectral library were then used for the targeted data extraction of the WiSIM-DIA data. The extracted ion chromatograms of the 12C and 13C isotopes for the precursor ions of each targeted peptide were used for quantification with a ±5 ppm window. The eight most-intense fragment ions (b and y types larger than 200 m/z) detected from the discovery data were used for peptide sequence confirmation through spectral library match within the 12 m/z CID MS-MS spectra.
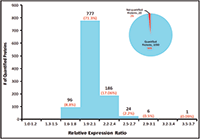
Figure 5: Quantification summary for quantifying 1100 E. coli proteins.
The relative expression ratios of targeted proteins between the 250 ng E. coli digest and 500 ng E. coli digest and CVs of each sample were calculated automatically. The p-value for each targeted peptide was also determined based on the spectrum match result. In this example, 5923 targeted E. coli peptides were identified with p-values less than 0.1 and %CVs ≤25% and were used to determine the relative expression ratios between samples. These 5923 identified peptides represent 1090 E. coli proteins yielding a 98% success rate for quantifying a total of 1100 targeted proteins (Figure 6). More than 97% of quantified proteins gave exceptional quantitative accuracy for the detection of the twofold expression change expected between the two samples (Figure 5). In addition, 85% of the 5923 quantified peptides gave %CVs less than 15% (Figure 6).
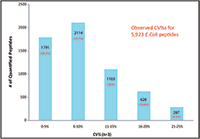
Figure 6: Coefficient of variation of 5923 quantified E. coli peptides.
Conclusion
A unique data-independent acquisition workflow that collects HRAM wide-window SIM data and rapid CID MS-MS data in parallel on the recent architecture MS, which combines a mass resolving quadrupole, an orbital trap mass analyzer, and a linear ion trap mass analyzer was developed. The method setup is simple, generic, and applicable to different types of samples. Any precursor ions detected by HRAM SIM acquisition can be quantified using an extracted ion chromatogram with a ±5 ppm window and simultaneously confirmed using DIA-CID MS-MS by applying a targeted data extraction approach post acquisition.
By quantifying using high-resolution (240,000) data that provides higher mass accuracy for separating precursor ions from background interferences, low limits of detection and high selectivity were achieved. We observed 10 amol on-column LOD and LOQ and four orders of linear dynamic range with the developed DIA workflow. The reproducible qualitative and quantitative record for each sample allowed large-scale quantification of more than 1000 proteins of interest with excellent analytical precision and great quantitative accuracy.
Reiko Kiyonami, Michael Senko, Vlad Zabrouskov, and Andreas F.R. Hühmer are with Thermo Fisher Scientific in San Jose, California. Jarrett Egertson, Sonia Ting, and Michael MacCoss are with the University of Washington in Seattle, Washington. Direct correspondence to: reiko.kiyonami@thermofisher.com
References
(1) L.C. Gillet, P. Navarro, S. Tate, H. Roest, N. Selevsek, L. Reiter, R. Bonner, and R. Aebersold, Mol. Cell Proteomics 11(6), O111.016717 (2012).
(2) J.D. Egertson, A. Kuehn, G. Merrihew, N. Bateman, B. MacLean, Y.S. Ting, J.D. Canterbury, M. Kellmann, V. Zabrouskov, C. Wu, and M.J. MacCoss, Nat. Methods 10, 744–746 (2013)
(3) S. Gallien, E. Duriez, K. Demeure, and B. Domon, J. Proteomics 81, 148–158 (2013).
(4) A. Prakash, D.M. Tomazela, B. Frewen, B. Maclean, G. Merrihew, S.M. Peterman, and M.J. MacCoss, J. Proteome Res. 8, 2733–2739 (2009).

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
Innovative Drug Delivery System Enhances Rhein's Bioavailability and Therapeutic Potential
January 16th 2025Researchers from Northern Border University in Saudi Arabia have developed a rhein-loaded self-nano emulsifying drug delivery system (RS-SNEDDS), significantly enhancing the compound's solubility, bioavailability, and therapeutic potential, particularly for neurological applications.