Managing Mass Spectrometric Data
Aspects of integrated data management specifically for mass spectrometric data are explored, including spectral dataset size, laboratory documents, and requirements for archiving and sharing research data.

Integrated data management is a central element of industrial research and manufacturing. However, data management within the university research setting is less well-defined. Variation in record keeping practices may be caused by the range of experience of researchers (from new undergraduate students to postdoctoral research associates), the exploratory nature of the research, and also perhaps a lack of consensus within the community in setting reasonable expectations. Here, we explore some aspects of integrated data management specifically for mass spectrometric data, touching on issues such as ever-increasing spectral dataset size, the breadth and complexity of associated laboratory documents, and evolving publication and funding requirements for archiving and sharing research data.
During lunch at a recent scientific conference, more folks at the table were checking their smart phones than eating their lunch. For those of us struggling to maintain some small part of the art of conversation, a science trivia question came up, at which point one of the "linked-in" generation diverted attention long enough to search for the answer on the web. Impressed, but trying to maintain the conversational thread, I ventured the opinion that the phones at the table had more interconnectivity and processing and data storage capability than the mass spectrometry (MS) data systems that I used at the start of my research career. Self-deprecation sparked interest from the younglings, and after explaining to the others what a Winchester disk drive was (1), I learned that the astrophysicist at the table simulated stellar accretion discs in a data file size of 1–10 terabytes, and that the protein sequencing scientist generated the same amount of data each and every month.
An Abundance of MS Data
Like any other modern instrument-based approach, MS analysis generates tremendous amounts of data. It is simple enough to examine the size of the files in our computers, or to examine storage requirements for archived measurements, and deduce that there has been a substantial increase in mass spectral dataset size over the last few decades. The number of experimental options also has increased. In the past, we may have focused on a single mass spectrum as the final result. Each compound was expected to have a distinct mass spectrum, usually an electron ionization mass spectrum recorded under standard conditions, and these spectra were gathered together in libraries (first printed, and now electronic) to serve as an archive of the spectral fingerprints used for compound identification. In the 1970s, it was gas chromatography coupled with mass spectrometry (GC–MS), and the associated necessity for measurement data in a computer for storage and processing, that represented the first of a series of major shifts in our approach. A GC–MS data file typically contains thousands of individual mass spectra, recorded sequentially. Spectra that span the time of an eluted peak could be averaged together, background-subtracted, and otherwise enhanced to produce the "fingerprint" mass spectrum that identifies the compound, or, given the synergism derived from over-determined data, provide clues about unresolved coeluted compounds. Fast forward 40 years, and consider all of the experimental options for recording mass spectra. The mass spectrum could be composed of positive ions, or negative ions, or possibly an alternating sequence. We might record low-resolution as well as high-resolution mass spectra. We might record product ion MS-MS spectra from any of several higher abundance ions in any of those mass spectra (recording those on the fly), or we might choose precursor ion or constant neutral loss MS-MS spectra. For certainty, we may elect to record the mass spectrum of a single compound with several different ionization methods or even with several different reagent gases in chemical ionization alone. We might derivatize the sample, age the sample, purify and extract and concentrate the sample, and complete another analysis. New instrumentation begets new chemistry; new insight begets new information.
With this array of measurement options, an associated increase in size and complexity of mass spectral datasets is inevitable. Figure 1 summarizes the increase in MS dataset size over the last few decades and forecasts possible continued growth into the future with new and broader applications of MS. The first three chevrons document 30 years from 1980 to the present. Amazingly, this period roughly encompasses the first widespread use of data systems for GC–MS (and MS dataset sizes of 20–40 megabytes) through to the modern laboratory that generates 1–10 terabytes of data monthly, including MS-MS and high-resolution mass spectral (HRMS) measurements. It is unrealistic to assume that such expansive sets of data can be interpreted and assessed solely through direct human effort. Figure 1 includes a break in the chronological sequence to emphasize the need for data mining, a generic term to include all of the processes in computer-assistance software to deal with large dataset sizes, perform certification and assurance tests, and to search for features and results, both apparent and hidden. Our ability to record massive amounts of data is a genuine challenge to automated procedures for assessment of that data, and as we will discuss in this column, managing it. The sequence outlined in Figure 1 also forecasts potential growth in MS datasets in the future. By 2020, for example, it is probable that sequencing datasets may expand to the petabyte threshold, especially as several sequencing experiments are linked together. Metabolomics analysis through MS includes complex experiments for hundreds of different metabolites. The analysis includes the use of isotopic tracers, with concordant control experiments, quantitation, and time-dependent analyses. Therefore, it is probable that by 2030, such uses of MS may reach the exabyte limit for summed dataset size.

Figure 1: Progression in the sizes of mass spectral datasets, linked to associated uses and decades.
Managing Data at the Petabyte Level and Beyond
Managing all of this data, even in consideration of the dataset size and scale alone, is clearly a challenging task. However, methods and strategies developed for comprehensive data management in scientific research, including MS (2,3), can be applied to this MS challenge. As in other research areas, we know that there is a substantial data record associated with mass spectral data that is not MS data itself, and proper data management methods encompass these records. For instance, the broader set of documents includes laboratory notebooks, instrument logs, sample inventories, blank files, calibration files, certification files, and — in the ideal case — link files to analytical data obtained on other mass spectrometers and other analytical instruments. In any data storage and management system, the format of the data becomes an important issue. That topic alone could be the focus of an entire column in this series, including formats that have been developed in efforts to reduce the size of the dataset. Martens and colleagues (4) recently described a community standard for MS data called mzML, developed from the Proteomics Standards Initiative. This user-developed initiative, developed only for the data, and covering only a portion of the associated metadata, is a hint at larger potentials that will greatly impact MS in the next 20 years. For example, the Institute for the Future (Palo Alto, California) states in their recent report (5):
Indeed, science has always been driven by data but now the sheer complexity and amount of information is demanding new practices and methods to translate the bits into knowledge. To that end, science will seek contributions from the networked public to tag raw data and make connections, seek patterns, and draw links between datasets. The speed of scientific discovery will be proportional to the availability of cross-disciplinary engagement with relevant data.
Although this is a forecast, the reality is already upon us (6).
Management Plan and Data Sharing
Reality in our scientific development derives from vision, realistic capabilities, and sometimes regulation and requirement. The latter (ideally at least) derives from a consensus understanding of what scientists do and how they do it. For this column, that latter desideratum is what mass spectrometrists do and how they do it. Figure 2 illustrates a data management timeline (applicable for any progression of scientific research), tracking from conception of the idea through to dissemination of data after publication of the results. Responsibilities for data management are plotted on the y-axis versus a time-related x-axis (note that this axis is nonlinear and has a break). There are issues of intellectual property, responsible mentoring, and community responsibilities all enfolded within the summed "data management" value, and many of these impact the university research environment. Research students, postdoctoral research associates, and university faculty working together to develop an idea and reduce it to practice all have interests in that intellectual property, and perhaps a patent application. There are specific record-keeping and data management practices to be followed.
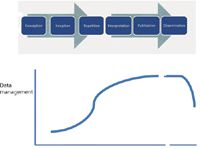
Figure 2: Data management responsibilities vary as research progresses from conception of experiment to dissemination of results.
The research progression in Figure 2 is marked out in six steps: conception, inception, repetition, interpretation, publication, and dissemination. Conception is, of course, the "What if?" and "How about if we?" creative discussions at the beginning of new research efforts. Because many ideas don't work out, it may seem as though there is no need for data management, but there is. Those first discussions may become important documents of the chronology later when claims of priority arise. Certainly, as ideas are put into practice (inception), responsibilities for data management increase, as first experiments provide some information about what works and what does not. First experiments may be difficult to describe completely and accurately because all of the relevant variables have not yet been identified. But then that's why they call it research. When the MS researcher gets a desired result, and wants to repeat it, the data management responsibility rises on the steepest part of the curve. Reproducibility by other researchers is a core value of scientific progress, and accurate experimental and instrumental records are vital. Interpretation of results (the next step) also relies on those records, and publication (the penultimate step) may require the submission of associated data as supplementary files to accompany the publication. In a future column, we will survey electronic records policies for MS data for the various MS journals.
The progression illustrated in Figure 2 is the philosophical overview of the evolution of an idea, but it also might be used to chart the progress of a graduate student's research efforts leading (ideally) to a dissertation. A research publication or the successful completion of a dissertation represents the culmination of an effort. Data management responsibilities in the last step of dissemination continue, at least for the period that the data is seen as current and relevant to others. Data have a half-life of usefulness, and only a limited subset of mass spectral data is valuable enough to be archived indefinitely. But in the period immediately after publication of a new research result, colleagues may wish to examine supporting data. They may have questions, seek additional data, or have differing interpretations of the data. After acceptance of a dissertation, and student graduation, those who follow in a research laboratory may depend on good data records to continue and extend research.
Therefore, there's an explicit expectation that scientific data and MS data will be shared within the community, and that expectation requires that records be kept and the data appropriately managed. Two related questions come to mind. First, if sharing is the expectation, then what's a reasonable time for maintaining records so that the data can be shared? (And what's the reason for the drop on the right side of the Figure 2 curve?) Second, who owns the data?
How long are data relevant? The answer to this question varies by research field and type of data. Survey data, for example, would reasonably have a greater lifetime than spectral analysis data, because surveys are seldom exactly repeated and are compared with each other in a trending analysis. In instrumental analysis, the finite lifetime of instruments factors into the archival value of the data produced. We can estimate the useful life of mass spectral data by examining aggregate descriptors for mass spectral publications, most of which are associated with supporting spectral data. The commercial service Journal Citation Reports compiles metrics on science journals, including a metric called "journal cited half life." For any given publication year, this factor tallies all citations to publications in that journal. The half-life is the point at which 50% of citations are to more recent publications in that journal, and 50% to older. "Rapid communications" journals are expected to have a shorter half-life factor than review journals, but the values calculated for MS journals and the range informs the related question of how long data are relevant to our own community. In a nutshell, the half-life for MS data is about five years.
The estimated half-life of five years helps to explain the drop off in the curve in Figure 2 after publication of mass spectral data. In a university research laboratory with a constant turnover of students and instruments, five years is also a reasonable estimate of data persistence. But a more prosaic reason contributes to the drop — regulations regarding the retention of research data. University policy may vary or may not exist at all. However, funding agencies impose requirements as a condition of receipt of research funding. As an example, the National Science Foundation (NSF, Arlington, Virginia) (7) requires that records (including data) pertinent to a grant be retained for three years from the date of submission of the "Final Report" for the award. Additionally, "NSF may request transfer to its custody of records not needed by the grantee when it determines that the records possess long-term retention value."
Now on to the second question: Who owns the data? In federally-funded research at universities, the answer is clear. But interestingly, the responses from those who perform the research are not consistent with the correct answer. Most principal investigators on federal awards claim that they own the data; some of the postdoctoral research associates or graduate students who complete the research, perhaps as part of a dissertation, assert that they own the data. However, the correct answer is that the grantee owns the data; the grantee is the university to whom the research funds were awarded. It is the grantee that is responsible for the proper administration of the award, and proper administration includes compliance with data sharing requirements. For instance, with respect to research awards from NSF (8):
Investigators are expected to share with other researchers, at no more than incremental cost and within a reasonable time, the primary data, samples, physical collections and other supporting materials created or gathered in the course of work under NSF grants. Grantees are expected to encourage and facilitate such sharing.
The National Institutes of Health (NIH, Bethesda, Maryland) has a specific data sharing policy that requires a data management plan for proposals that seek more than $500,000 in direct costs annually. Specific types of projects funded by NIH have individualized data-sharing requirements. For NSF (9):
Proposals submitted or due on or after January 18, 2011, must include a supplementary document of no more than two pages labeled "Data Management Plan." This supplementary document should describe how the proposal will conform to NSF policy on the dissemination and sharing of research results
As mass spectrometrists, we may tend to view our data as value-neutral, of interest to ourselves in analysis, method development, and creative research. But data are a valuable commodity and resource, and while it seems that there is an increasing modern emphasis on its archival and subsequent use, taking advantage of new computational capabilities, access to the data itself, and concerns about its quality, have been the subject of legislation in the United States for some time. In 1999, the "Shelby amendment" to an omnibus funding bill provided direction to the Office of Management and Budget (OMB) to take action so that the data that result from federally-funded research are available through Freedom-of-Information Act (FOIA) requests (10). The "Data Quality Act" (DQA) passed in 2001 as part of an appropriations bill; here the OMB was to create guidelines that "provide policy and procedural guidance to Federal agencies for ensuring and maximizing the quality, objectivity, utility, and integrity of information (including statistical information) disseminated by Federal agencies." Apparently, "quality" was a flashpoint term, with the DQA later coming under criticism for its lack of legislative depth and debate, and its potential for abuse (11,12). As this article was written, the U.S. Federal government issued a request for information "Public Access to Digital Data Resulting from Federally Funded Scientific Research" (13). Although the public comment period for this request ended January 12, 2012, a future Federal Register will include the public comments received. The request for information contains 13 insightful questions dealing with issues of data formats, competing interests of stakeholders, intellectual property, international communities, and the potential role of data standards. Interestingly, the issue of increasing dataset size is not represented within the list of questions, nor are questions presented that discuss physical aspects (cost and location) of any potential centralized data archive. Even as we have created data archives, the cost to maintain them has become problematic. In spring 2011, NIH announced plans to shutter bioinformatics databases, including the Sequence Reach Archive repository, because of budget restrictions. A company with some experience in large databases made a $15-million grant to maintain accessibility in a nongovernment hosting site. Thank you, Google (14).
Conclusion
In the end, intellectual property concerns associated with data management and data sharing are likely to be the most challenging, and the most engaging. Must investigators share their datasets with their colleagues when they view their colleagues as competitors for research funding? What if special software or algorithms are needed to access or process the data, and those are under separate patent or proprietary protections? Data are indeed a treasure trove; after the mine is located, will data wildcatters appear to siphon value? Legislation has been passed, and guidelines put in place, for data management and data sharing. The assessment of reasonableness, driven in no small part by community consensus, lags behind. But we are moving inexorably into the large scale data universe, whether we are ready for it or not.
Kenneth L. Busch fearlessly forecasts MS dataset size 30 years into the future in this column. He is convinced that if it were not for the fact that mass spectral data has a relatively short half-life, we would be buried under paper copies of billions of mass spectra previously recorded. He concludes that most of the mass spectral data archived today was probably recorded within the past five years, and that newer data will overwhelm the archives unless explicit and reasonable efforts are taken to purge older data. He invites readers to check back with him in 2038 to see if his dataset size forecast for 2040 is on track. This column is the sole responsibility of the author, who can be reached at WyvernAssoc@yahoo.com.

Kenneth L. Busch
References
(1) http://en.wikipedia.org/wiki/History_of_hard_disk_drives.
(2) E.E. Schadt, M.D. Linderman, J. Sorenson, L. Lee, and G.P. Nolan, Nat.Rev. Genet. 11, 647–657 (2010).
(3) T. Kosar, Data Intensive Distributed Computing: Challenges and Solutions for Large-Scale Information Management, (IGI Global, 2012; ISBN-10: 1615209719).
(4) L. Martens, M. Chambers, M. Sturm, D. Kessner, F. Levander, J. Shofstahl, W.H. Tang, A. Rompp, S. Neumann, A.D. Pizarro, L. Montecchi-Palazzi, N. Tasman, M. Coleman, F. Reisinger, P. Souda, H. Hermjakob, P.-A. Binz, and E.W. Deutsch, Mol. Cel. Proteom. 10: 10.1074/mcp.R110.000133, 1–7 (2011).
(5) "A Multiverse of Exploration: The Future of Science 2021," Institute for the Future, Technology Horizons, Fall 2011.
(6) http://www.pnas.org/content/early/2011/11/02/1115898108.
(7) "NSF Proposal and Award Policies and Procedures Guide," NSF 09-29, Section II-9.
(8) "NSF Proposal and Award Policies and Procedures Guide," NSF 09-29, Section VI-8b.
(9) http://www.nsf.gov/bfa/dias/policy/dmp.jsp.
(10) http://www.aaas.org/spp/cstc/briefs/accesstodata/index.shtml.
(11) C. Mooney, The Republican War on Science (Basic Books, New York, 2005), p. 103.
(12) C.C. Meshkin, Engage 11(3), 44 (2010).
(13) Federal Register, 76(214), 68518–68520, FR Doc No: 2011-28623 (2011).
(14) R. Dragani, Biotechniq. 51(5), (2011) http://www.biotechniques.com/news/Google-to-the-Rescue/biotechniques-323607.html.

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.

.png&w=3840&q=75)

.png&w=3840&q=75)



.png&w=3840&q=75)



.png&w=3840&q=75)










