Resonance Light Scattering Spectrum of the Alizarin Green-CTMAB-Nucleic Acids System and Determination of Nucleic Acids at Nanogram Levels
Spectroscopy
The study of the interaction mechanism of nucleic acids-CTMAB with AG showed that the enhanced RLS comes from the aggregation of AG on nucleic acids through the bridged and synergistic effect of CTMAB.

Investigations of the interaction of small molecules with nucleic acids have been the focus of extensive research in recent years. The quantitative determination of nucleic acids is the basis of many studies. But direct use of the intrinsic fluorescence and ultraviolet absorption of nucleic acids for the determination has been severely limited by low sensitivity and serious interference (1–3).
Rayleigh scattering is a light-scattering phenomenon in which electromagnetic radiation and material act on each other and generate elastic collisions. According to the macroscopic fluctuation theory, the Rayleigh scattering law is obeyed (4) for the light scattering of the molecular particles 20-fold smaller than the wavelength of the incident beam. However, if the wavelength of the incident beam is close to that of the absorption band of the molecular particles, which exit as aggregates, Rayleigh scattering will deviate from the law and the intensity of some wavelengths will increase rapidly. This phenomenon is called resonance light scattering, or resonance-enhanced Rayleigh scattering, or resonance Rayleigh scattering (5). By using a common spectrofluorometer to measure the RLS, Pasternake and colleagues (6,7) studied the aggregations of porphyrins. Later, the RLS technique was used for determining nucleic acid and protein (8,9).
The probes for nucleic acids include cation porphyrins (6,10), triphenylmethane dyes (11,12), phenothiazinium dyes (13,14), xanthene dyes, (15) metal complexes, (16,17) and nanoparticles (18,19). However, few investigations have been carried out to apply the anionic dyes as probe reagents to determine nucleic acids so far, due to the electrorepulsion between nucleic acids and anionic dyes.
By introducing the cationic surfactant CTMAB into the nucleic acids–anionic dyes system, we noticed that DNA, CTMAB, and anionic dyes can form a three-component complex, which results in enhanced RLS signals and can be applied successfully to determine DNA by the RLS method. At pH 6.75 and ionic strength 0.07, the anionic dye alizarin green (AG, Figure 1, displaying its molecular structure) can aggregate on the surface of DNA through the bridged and synergistic effect of CTMAB, which results in the enhanced IRLS of AG-CTMAB-yDNA system. The reaction mechanism and the influential factors on the variation of RLS have been investigated tentatively. This method enlarges the range of probe reagents of nucleic acids, which will be applied widely for quantitative determination of nucleic acids.
Figure 1
Experimental
Chemicals
All reagents were of analytical reagent grade without further purification, and doubly distilled water was used throughout.
Stock solutions of nucleic acids were prepared by dissolving commercially purchased yeast DNA (Shanghai Changyan Pharmaceutical factory, Shanghai, China), calf thymus DNA (Shanghai Changyan Pharmaceutical factory), fish sperm DNA (fsDNA, Shanghai Institute of Biochemistry, Academy of Science, Shanghai, China), and yeast RNA (Shanghai Changyan Pharmaceutical factory) in doubly distilled water. These stocks needed to be stored at 0–4 °C with only an occasional gentle shake if needed. The concentration of DNA is determined according to the absorbance values at 260 nm by using εDNA = 6600 mol–1 l cm–1 and εRNA = 7800 mol–1 l cm–1 , respectively (20). In this experiment, all working solutions of nucleic acids were 25.0 mg/L.
5.0 × 10–4 mol/L stock solution of alizarin green (AG, Shanghai Chemical Reagents Co., Shanghai, China), was prepared by dissolving 0.3111 g of the crystal product in water in a 1000-mL volumetric flask.
A 5.0 × 10–4 mol/L stock solution of cation surfactant cetyltrimethylammonium bromide (CTMAB, Merck, Germany), was prepared by dissolving 0.1832 g of the crystal product in water in a 1000-mL volumetric flask.
A 0.10-mol/L HMTA-HCl buffer solution was prepared by dissolving 0.1401 g hexamethylene tetramine (HMTA) in a 1000-mL volumetric flask in water, and adjusting the pH to 6.75 with HCl.
Apparatus
The RLS spectrum and the intensity of RLS were measured with an RF-5301PC (Shimadzu, Japan) fluorescence spectrometer by using a 1.00-cm quartz fluorescence cell. All absorption spectra were measured on a UV-265 spectrophotometer (Shimadzu, Japan). The surface tension (σ) was measured with a KrÜss KizMK5 programe surface tension instrument (Krss GmbH, Hamburg, Germany) by using the suspended plate method. A pHs-3C digital pH meter (Leici, Shanghai) was utilized to detect the pH values of the aqueous solutions.
Experimental Procedure
Into a 25-mL volumetric test tube was added an appropriate volume of DNA solution according to the concentration needed, 0.50 mL 5.0 × 10–4 mol/L CTMAB, 0.50 mL 5.0 × 10–4 mol/L AG, and 2.00 mL HMTA-HCl buffer. The mixture was diluted to the mark with water and mixed thoroughly, and was allowed to stand still for 10 min before RLS measurements. All measurements were made at an ambient temperature of 25 °C.
All RLS spectra were obtained by scanning the excitation and emission monochromators simultaneously (namely, Δλ = 0 nm) from 250.0 to 700.0 nm. The intensity of RLS was measured at λ = 340.0 nm in a quartz fluorescence cell with slit width at 3.0 nm for the excitation and emission. The enhanced intensity of the CTMAB-AG system by DNA was represented as ΔI RLS = I RLS – I0 RLS . Here, I RLS and I0 RLS were the intensity of the system with and without DNA.
Results and Discussion
Features of the Resonance Light Scattering Spectrum
Figure 2 (curves 1–6) shows the light-scattering spectra of AG-CTMAB-yDNA-buffer, yDNA-CTMAB-buffer, AG-yDNA-buffer, AG-buffer, AG, and CTMAB-AG-buffer. It can be seen from the figure that their RLS intensities exhibit different features. In the whole scanning region, the RLS intensities of curve 2 and curve 3 are stronger than those of curve 6, but are still much weaker than those of curve 1, illustrating that the RLS of DNA is obviously enhanced with the help of both AG and CTMAB. Curve 4 and curve 5 are almost the same, which shows that the effect on the ΔI RLS values of adding HTMA to AG is weak. Similar RLS spectral characteristics can be surveyed when fsDNA, ctDNA, and RNA are used.
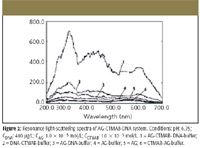
Figure 2
yDNA is a sort of surface-inactive anionic polyelectrolyte, and AG is also in the anionic state at pH 6.75. The electrorepulsion force between AG and DNA will prevent them from reacting with each other. CTMAB is a cationic surfactant; it can react with both AG and DNA due to electrostatic force to produce a kind of three-component complex (21) of AG-CTMAB-DNA at pH 6.75. The CTMAB in the system acts as a bridge between AG, and DNA and has a synergistic effect, which results in the greatly enhanced RLS intensities of DNA.
Because the enhanced extents of the RLS of AG-CTMAB-DNA system at 340.0 nm are in good proportion to the concentration of DNA, the 340.0 nm wavelength is selected for further research.

Figure 3
Effect of Buffer and Surfactant
The effects of surfactant are examined and shown in Figure 3. Experiment results prove that different kinds of surfactants had different effects on ΔI RLS , with CTMAB being the best surfactant. The effects of different buffers on ΔI RLS of AG-CTMAB-DNA system are investigated, and HMTA is the best buffer tested. The effects of CTMAB and pH are shown in Figure 4 and Figure 6, respectively. In Figure 4, ΔI RLS increases with increasing the concentration of CTMAB when it is below 7.5 × 10–6 mol/L because CTMAB can form an ionic association complex with the phosphate groups of nucleic acids at appropriate concentration. Thus, the volume or size of nucleic acids acting as a template that AG assembles on is enlarged, which results in the enhanced ΔI RLS of the system. The ΔI RLS decreases when the concentration is above 3.0 × 10–5 mol/L and is constant in the range 7.5 × 10–6 to 3.0 × 10–5 mol/L.
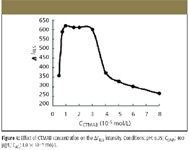
Figure 4
From the measurement of surface tension (Figure 5), we can obtain the critical associate concentration (CAC) in this system, which is 1.0 × 10–5 mol/L, so 1.0 × 10–5 mol/L CTMAB is selected for further research. Figure 6 shows that the enhancement of light scattering changes a lot and reaches a maximum with pH in the range 6.00 to 7.00. The dependence of light scattering upon pH might be relevant to the form of AG and DNA; when pH is below 6.00, both the free base form of AG and DNA are protonized, which is not favorable for positively charged CTMAB to be close to the DNA molecule. At the same time, the concentration of AG with two negative charges is low, which is not favorable for the reaction of AG and CTMAB. When the pH is above 7.00, the ΔI RLS of AG markedly decreases. So pH 6.75 is chosen for further research.
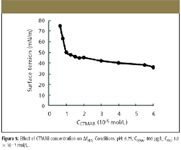
Figure 5
Effect of Alizarin Green Concentration
The effect of alizarin green concentration on ΔI RLS of the AG-CTMAB-DNA system is investigated and shown in Figure 7. When the concentration of alizarin green is in the range 8.0 × 10–7 to 1.6 × 10–6 mol/L, the ΔI RLS of the AG-CTMAB-DNA system reaches a maximum. So we select 1.0 × 10–6 mol/L AG for further research.
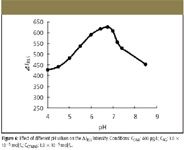
Figure 6
Effect of Addition Order of the Reagents
It is very important that the addition order of reagents be fixed. Our experiments show the optimal addition order of the reagents to be as follows: DNA should be mixed with CTMAB first, and then AG and the buffer can be added in. When AG, CTMAB, DNA, and buffer are added successively, the ΔI RLS of the system is much smaller (6). The study of the interaction mechanism of nucleic acids-CTMAB with AG shows that changing the order of reagents can alter the initial concentrations of DNA. When CTMAB reacts with DNA first, this is favorable for the aggregation of AG on the surface of the DNA molecule.
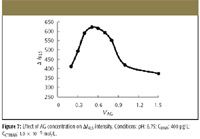
Figure 7
Effect of the Ionic Strength
The effect of ionic strength on ΔI RLS of the AG-CTMAB-DNA system is shown in Figure 8. The ΔI RLS of the AG-CTMAB-DNA system lessens markedly with the increasing ionic strength, showing that electrostatic makes a key contribution to the interaction of CTMAB, DNA, and AG. Hydrophobic force probably does its work in the creation of the three-component complex of AG-CTMAB-DNA.

Figure 8
The complex of AG-CTMAB-DNA is sensitive to ionic strength of medium, and ΔI RLS decreases with the increasing ionic strength due to the weakening combination of the complex but changes slightly when the ionic strength is below 0.07 mol/L. So to get the best ΔI RLS in this work, ionic strength must be controlled to be below 0.07 mol/L.
Reaction Time and Stability
The binding reaction of DNA and AG with CTMAB is completed after 10 min. They are mixed together at room temperature (25 °C); ΔI RLS remains a constant for about 3 h. The reaction has good stability.
Tolerance of Foreign Coexisting Substances
The effect of substances including metal ions, amino acids, galactose, and adenine are examined for interference. The results are summarized in Table I. Most of the metal ions in biological systems, such as K+ , Na+ , and Ca2+ , can be tolerated at high concentrations.
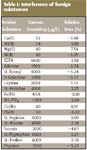
Table I: Interference of foreign substances
Analytical Application
Calibration
Under optimum conditions, the dependence of ΔI RLS upon the concentration of DNA is determined. The analytical parameters of this method are listed in Table II. Table II also shows that there was a good linear relationship between ΔI RLS and DNA in a wide range.
No groove and helix structure exists in RNA; however, both DNA and RNA can increase the RLS intensity of AG-CTMAB in our experiment. So groove binding and intercalative binding are not the reason for RLS enhancement.
Determination of DNA in Synthetic Samples
With the calibration curves, two synthetic samples constructed on the basis of the interference of foreign coexisting substances are determined simultaneously under the same conditions. Their determination results are listed in Table III. Table III reveals that the values found in synthetic samples are identical to the expected ones; the recovery is satisfactory.

Table II: Analytical parameters for the determination of nucleic acids
Study on the Interaction Between Alizarin Green, CTMAB, and DNA
Generally, organic dyes interact with DNA in the following models: intercalative, groove, or electrostatic binding, and the long-range assembly on the molecular surface of DNA does not involve intercalative or groove binding. The addition of CTMAB and DNA can enhance the ΔI RLS without AG (Figure 2), which is due to the formation of a DNA-CTMAB association, although the ΔI RLS is smaller than that in the presence of AG. AG and DNA cannot react with each other due to the electrostatic between them (23). By introducing a bridge — namely, the cationic surfactant CTMAB — into a DNA-anionic dyes system, AG, CTMAB, and DNA formed a three-component complex of AG-DNA-CTMAB, which shortened the distance between AG and DNA and enhanced the assembly of AG on the surface of DNA.

Table III: Determination of DNA in synthetic sample
Several other surfactants, including TPB, Tritonx-100, SDBS, and SDS (their concentrations are all in 1.0 × 10–6 mol/L range) have been employed to examine further the surfactant effects functioning in RLS enhancement. Taking the interacting system of AG-CTMAB-yDNA system as an example, the percentages of ΔI RLS are 80.3%, 28.4%, 10.6%, and 8.93% for TPB, Triton x-100, SDBS, and SDS, respectively (assuming that the percentage of ΔI RLS of AG-CTMAB-yDNA is 100%). Apparently, cation surfactants reveal RLS enhancement effects much more prominently than both non-ion surfactants and anion surfactants, disclosing that electrostatic force performs an important function in the combination of DNA and AG with surfactant. At the same time, the relatively weak RLS enhancement effects of non-ion and anion surfactant confirm that hydrophobic force is also a factor in the reaction of DNA and AG with surfactant (24).
Viscosity is one of the important parameters to assess the binding mode of a molecule to DNA (25,26). The relative viscosities of different mixtures were examined in this experiment and the results shown in Table IV. The relative viscosity of DNA declined with the addition of CTMAB, which showed that there was no intercalative between CTMAB and DNA, as well as the system of AG-CTMAB-DNA.

Table IV: Determination result of relative viscosity
To understand the interaction mechanism, it is important to observe the molecular absorption features of the AG-CTMAB-yDNA complex. According to the theory of RLS (4,7,27), the RLS peak at 340 nm is ascribed to the molecular absorption band of the system in the range 260–300 nm (Figure 2), and the RLS peak at 409 nm and 544 nm to the absorption band at 415 nm and the range 580–660 nm of the system. The light absorption of the system (Figure 9) increases with increasing the concentration of DNA until CDNA is 2.5 × 10–6 g/mL, because of the forming of large assembly products. However, if the concentration of DNA keeps increasing, the maximum absorption of AG-CTMAB-yDNA complex decreases. The curves show that there is almost no bathochromic or hypsochromic shifts that observe the intercalation of CTMAB and AG with DNA, because the bathochromic and hypochromic effects in the absorption spectra are associated with the intercalation of small molecules into the adjacent base pairs of DNA (28). Similar phenomena also were found for the interaction of AG with ctDNA, fsDNA, and RNA, which disclosed further that the intercalation of CTMAB and AG with DNA is not groove binding and intercalative binding.

Figure 9
Herein, the interaction of DNA, CTMAB, and AG mainly depends upon electrostatic binding. Hydrophobic force is also a factor in the reaction of DNA and AG with surfactant.
Conclusion
The nature of RLS from an aqueous solution of the ion-association complex of nucleic acids–CTMAB-AG system was studied for the first time. The relationship between ΔI RLS and the concentration of nucleic acids was established. A new way to determine nonagram levels of nucleic acids by the RLS spectra by using a common spectrofluorometer was proposed. The study of the interaction mechanism of nucleic acids–CTMAB with AG showed that CTMAB can interact with AG and nucleic acids and form a three-component complex, which results in the enhanced RLS signals and can be applied successfully to determine DNA by RLS method. This method has the following advantages: the incubation time is short; it has high selectivity and simplicity of operation; it enlarges the range of probe reagents of nucleic acids; and it will be applied widely for quantitative determination of nucleic acids in the chemical and biochemical fields.
Acknowledgments
This work was supported by the Provincial Science Foundation of Hunan Province, People's Republic of China (00JJY2084) and the Educational Department Science Foundation of Hunan Province (03c611), People's Republic of China.
Xioaming Chen, Changqun Cai, Hang Gong, and He'an Luo are with the Institute of Chemistry, Xiangtan University, Xiangtan, China.
References
(1) S. Udemfriend and P. Zaltaman, Anal. Biochem. 3, 49 (1962).
(2) W.E. Schy and M.J. Pleva, Anal. Biochem. 180, 314 (1989).
(3) Z.-X. Guo and H.-X. Shen, Analyst 124, 1093–1098 (1999).
(4) G.A. Miller, J. Phys. Chem. 82, 616 (1978).
(5) J. Aglister and I.Z. Steinberg, J. Chem. Phys. 78, 5358 (1983).
(6) C.Z. Huang, K.A. Li, and S.Y. Tong, Anal. Chem. 68, 2259–2263 (1996).
(7) R.F. Pasternack and P.J. Collings, Science 269, 935 (1995).
(8) C.Q. Ma, K.A. Li, and S.Y. Tong, Anal. Chim. Acta 338, 255 (1997).
(9) Y.J. Chen, J.H. Yang, X. Wu, T. Wu, and Y.X. Luan, Talanta 58, 869 (2002).
(10) Z.X. Guo, L. Li, H.X. Shen, and X. Cong, Anal. Chim. Acta 379, 45–51 (1999).
(11) C.Z. Huang, Y.F. Li, X.H. Huang, and M. Li, Anal. Lett. 34, 1117–1132 (2001).
(12) X.L. Chen, D.H. Li, Q.Z. Zhu, H.H. Yang, H. Zheng, and J.G. Xu, Chem. J. Chinese Univ. 22, 901 (2001).
(13) Y.T. Wang, F.L. Zhao, K.A. Li, and S.Y. Tong, Anal. Chim. Acta 396, 75–81 (1999).
(14) Y. Liu, C.Q. Ma, K.A. Li, and S.Y. Tong, Anal. Biochem. 268, 187–192 (1999).
(15) H.L. Wu, W.Y. Li, X.W. He, and H. Liang, J. Anal. Sci. 18, 94 (2002).
(16) Y.M. Hao and H.X. Shen, Anal. Chim. Acta 413, 87–94 (2000).
(17) Y.M. Hao and H.X. Shen, Anal. Chim.Acta 422, 159–166 (2000).
(18) J. Yguerabide, E.E. Yguerabide, G. Bee, K. Yamout, L. Korb, J. Beck, and T. Peterson, Nat. Gen. 23, 67–68 (1999).
(19) P. Bao, A.G. Frutos, C. Greef, J. Lahiri, U. Muller, T.C. Peterson, L. Warden, and X.Y. Xie, Anal. Chem. 74, 1792–1797 (2002).
(20) Z. Chen, J. Liu, and D. Luo, "Biochemistry Experiments", Chinese University of Sciences and Technology Press, Hefei, China (1994).
(21) R.T. Liu, J.H. Yang, X. Wu, and T. Wu. Anal. Chim. Acta. 448, 85–91 (2001).
(22) Y.H. Zhang and Y.Q. Liang, Chem. J. Chin. Univ. 2, 266 (1994).

LIBS Illuminates the Hidden Health Risks of Indoor Welding and Soldering
April 23rd 2025A new dual-spectroscopy approach reveals real-time pollution threats in indoor workspaces. Chinese researchers have pioneered the use of laser-induced breakdown spectroscopy (LIBS) and aerosol mass spectrometry to uncover and monitor harmful heavy metal and dust emissions from soldering and welding in real-time. These complementary tools offer a fast, accurate means to evaluate air quality threats in industrial and indoor environments—where people spend most of their time.
NIR Spectroscopy Explored as Sustainable Approach to Detecting Bovine Mastitis
April 23rd 2025A new study published in Applied Food Research demonstrates that near-infrared spectroscopy (NIRS) can effectively detect subclinical bovine mastitis in milk, offering a fast, non-invasive method to guide targeted antibiotic treatment and support sustainable dairy practices.