The Use of Raman Spectroscopy in Cancer Diagnostics
Application Notebook
Both Raman spectroscopy and surface-enhanced Raman spectroscopy (SERS) are proving to be invaluable tools in the field of biomedical research and clinical diagnostics.
Both Raman spectroscopy and surface-enhanced Raman spectroscopy (SERS) are proving to be invaluable tools in the field of biomedical research and clinical diagnostics. The robust, compact, fit-for-purpose Raman spectrometer designs are appropriate for use in surgical procedures to help surgeons assess tumors and allow rapid decisions to be made. Raman systems are also being developed for molecular diagnostic testing to detect and measure human cancer biomarkers. Based on the SERS technique, this approach potentially could change the way bioassays are performed to improve both the sensitivity and reliability of testing. The two applications highlighted in this review, together with other examples of the use of Raman spectrometry in biomedical research areas such as the identification of bacterial infections, are clearly going to make the technique an important part of the medical toolbox, as we continually strive to improve diagnostic techniques and bring a better health care system to patients.
In recent years, Raman spectroscopy has gained widespread acceptance in applications that span from the rapid identification of unknown components to detailed characterization of materials and biological samples. The technique's breadth of application is too wide to reference here, but examples include quality control (QC) testing and verification of high purity chemicals and raw materials in the pharmaceuticals and food industries (1–3), investigation of counterfeit drugs (4), classification of polymers and plastics (5), characterization of tumors, and the detection of molecular biomarkers in disease diagnostics (6–8) and theranostics (9).
One of the most exciting application areas is in the biomedical sciences. The major reason behind this surge of interest is that Raman spectroscopy is an ideal technique for molecular fingerprinting and is sensitive to the chemical changes associated with disease. Furthermore, components in the tissue matrix, principally those associated with water and bodily fluids, give a weak Raman response, thereby improving sensitivity for those changes associated with a diseased state (10). The growing body of knowledge in our understanding of the science of disease diagnostics and improvements in the technology has led to the development of fit-for-purpose Raman systems designed for use in surgical theaters and doctors' surgeries, without the need for sending samples to the pathology laboratory (8).
Surface-enhanced Raman spectroscopy (SERS) is a more sensitive version of Raman spectroscopy, relying on the principle that Raman scattering is enhanced by several orders of magnitude when the sample is deposited onto a roughened metallic surface. The benefit of SERS for these types of applications is that it provides well-defined, distinct spectral information, enabling characterization of various states of a disease to be detected at much lower levels. Some of the many applications of Raman and SERS for biomedical monitoring include
- Examination of biopsy samples
- In vitro diagnostics
- Cytology investigations at the cellular level
- Bioassay measurements
- Histopathology using microscopy
- Direct investigation of cancerous tissues
- Surgical targets and treatment monitoring
- Deep tissue studies
- Drug efficacy studies
The basics of Raman spectroscopy are well covered elsewhere in the literature (11). However, before we present some typical examples of both Raman and SERS applications in the field of cancer diagnostics, let's take a closer look at the fundamental principles and advantages of SERS.
Principles of Surface-Enhanced Raman Spectroscopy
The principles of SERS are based on amplifying the Raman scattering using metal surfaces (usually Ag, Au, or Cu), which have a nanoscale roughness with features of dimensions 20–300 nm. The observed enhancement of up to 107-fold can be attributed to both chemical and electromagnetic effects. The chemical component is based on the formation of a charge-transfer state between the metal and the adsorbed scattering component in the sample and contributes about three orders of magnitude to the overall enhancement. The remainder of the signal improvement is generated by an electromagnetic effect from the collective oscillation of excited electrons that results when a metal is irradiated with light. This process, known as the surface plasmon resonance (SPR) effect, has a wavelength dependence related to the roughness and atomic structure of the metallic surfaces and the size and shape of the nanoparticles. For a more detailed discussion of SERS and its applications, see reference 12.
Detection by SERS has several benefits. Firstly, fluorescence is inherently quenched for an adsorbate analyte on a SERS surface resulting in fluorescence-free SERS spectra. This is primarily because the Raman signal is enhanced by the metal surface, and the fluorescence signal is not. As a result, the relative intensity of the fluorescence is significantly lower than the Raman signal and in many occasions is not observable. Secondly, signal averaging can be extended to increase sensitivity and lower detection levels. Finally, SERS spectral bands are very sharp and well-defined, which improves data quality, interpretability, and masking by interferents. Recent work using silver nanoparticles as the enhancing substrate has shown that SERS can be applied to single-molecule detection, rivaling the performance of fluorescence measurements (13).
Although SERS has many advantages it also has several disadvantages that are worth noting. The first is that unlike traditional Raman spectroscopy, SERS requires sample preparation. The ability to measure samples in vivo without the need for sample pretreatment is one of the major advantages of traditional Raman spectroscopy. For that reason, it is important to understand the signal enhancement benefits of SERS compared to the ease of sampling of traditional Raman spectroscopy when deciding which technique to use (14). Second, high performance SERS substrates are difficult to manufacture and, as a result, there is typically a high degree of spatial nonuniformity with respect to the signal intensity (15). For example, the SERS substrates used in the research being carried out by the University of Utah (described later in this article) tend to have much higher concentrations of analyte on the edges of the sampling area than in the center. Lastly, it is important to note that when the sample is deposited onto the surface, the molecular structure (the nature of the analyte) can be altered slightly resulting in differences between traditional Raman spectra and SERS spectra (16). However, it should be emphasized that there have been improvements in the quality of substrates being manufactured today and new materials such as Klarite (Renishaw Diagnostics) are showing a great deal of promise in terms of consistency and performance (17).
Raman Spectroscopy Applied to Biomedical Studies
There are several important functional groups related to biomedical testing that have characteristic Raman frequencies. Tissue samples include components such as lipids, fatty acids, and protein, all of which have vibrations in the Raman spectrum. The most significant spectral regions include
- X-H bonds (for example, C-H stretches): 4000–2500 cm-1 region
- Multiple bonds (such as N≡C): 2500–2000 cm-1 region
- Double bonds (for example, C=C, N=C): 2000–1500 cm-1 region
- Complex patterns (such as C-O, C-N, and bands in the fingerprint region): 1500–600 cm-1 region:
The identification and measurement of Raman scattering in these spectral regions can be applied to the following biomedical applications:
- Measurement of biological macromolecules such as nucleic acids, proteins, lipids, and fats
- Investigation of blood disorders such as anemia, leukemia, and thalassemias (inherited blood disorder)
- Detection and diagnosis of various cancers including brain, breast, pancreatic, cervical, and colon
- Assaying of biomarkers in studying human diseases
- Understanding cell growth in bacteria, phytoplankton, viruses, and other microorganisms
- Analysis of abnormalities in tissue samples such as brain, arteries, breast, bone, cervix, embryo media, esophagus, gastrointestinal tract, and the prostate gland
Some typical examples of Raman spectra of biological molecules are shown in Figure 1. Figure 2 gives a summary of some common biochemical functional groups with their molecular vibrational modes and frequency ranges, which are observed in compounds such as proteins, carbohydrates, fats, lipids, and amino acids (6,8,10,18–20). Identification of these functional groups can be used as a qualitative or quantitative assessment of a change or anomaly in a sample under investigation.
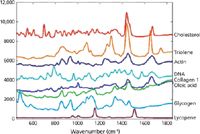
Figure 1: Raman spectra of various biological compounds. Adapted from reference 21.
Let's take a look at two examples of the use of both conventional Raman spectroscopy and SERS, specifically for cancer diagnostic purposes. The first example uses traditional Raman spectroscopy for the assessment of breast cancer and the second example takes a look at SERS for the screening of pancreatic cancer biomarkers.

Figure 2: Some common functional groups with their molecular vibrational modes and frequency ranges observed in various biochemical compounds. Adapted from reference 18.
Detection and Diagnosis of Breast Cancer
The surgical management of breast cancer for some patients involves two or more operations before a surgeon can be assured of removing all cancerous tissue. The first operation removes the visible breast cancer and samples the lymph nodes for signs of the disease spreading. Intraoperative methods for testing the health of the lymph nodes involve classical tissue mapping techniques such as touch imprint cytology and histopathological, frozen section microscopy. The problem with both of these methods is that they have poor sensitivity and require an experienced pathologist to interpret the data and relay the "benign" or "malignant" diagnostic report to the surgeon. For that reason, these intraoperative methods have not been widely adopted by the medical community and, as a result, most procedures still require samples to be sent to a laboratory for further testing. If tests confirm that the cancer has spread, then the patient must undergo an additional surgery to remove all the nodes in the affected area.
A preferable approach would be for an intraoperative assessment, which would allow for the immediate excision of all of the axillary lymph nodes, if required. Studies by researchers at Cranfield University and Gloucestershire Royal Hospital in the United Kingdom have shown that Raman spectroscopy can detect differences in tissue composition, particularly a wide range of cancer pathologies (22). This group used a portable Raman spectrometer with a 785-nm excitation laser wavelength and a fiber-optic probe (MiniRam, B&W Tek) and principal component analysis (PCA) along with linear discriminant analysis (LDA) data processing to evaluate the Raman spectra of lymph nodes. They confirmed that the technique can match the sensitivities and specificities of histopathological techniques that are currently being used.
The initial work in this study involved taking Raman spectra of the same samples that were being evaluated by the traditional intraoperative techniques. The study, which assessed 38 (25 negative and 13 positive) axillary lymph nodes from 20 patients undergoing surgery for newly diagnosed breast cancer, compared the results with a standard histopathological assessment of each node. The results showed a specificity of 100% and sensitivity of 92% when differentiating between normal and metastatic nodes. These results are an improvement on alternative intraoperative approaches and reduce the likelihood of false positive or false negative assessment (20). Typical Raman spectra of lymph nodes from benign breast tissue and malignant tumors are shown in Figure 3. It can be seen that while there are very subtle differences between the two spectra, the data classes can be discriminated using multivariate classification tools such as PCA-LDA and support vector machine (SVM) classification applied over the 800–1800 cm-1 spectral range. By using such supervised classification tools, Raman spectroscopy is sensitive enough to pick up the differences in the chemistry of the diseased state compared to that of the healthy tissue. For example, in Figure 3, subtle differences in the fatty acid peaks at 1300 and 1435 cm-1, and of collagen peaks at 1070 cm-1, are highlighted from the loadings plots from PCA analysis of the data (19).
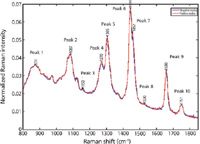
Figure 3: Raman spectral peaks of lymph nodes of benign breast tissue (blue), together with a malignant breast tumor (red). Adapted from reference 21.
More work is currently being done to support these findings at the University of Exeter and other research facilities. If confirmed, the next stage is to introduce the probe-based Raman spectrometer into an operating theater, where it can be used to assess the node as soon as it is removed from the patient.
Additionally, many other groups are working on different approaches to cancer screening using Raman spectroscopy. Recent studies at the Massachusetts Institute of Technology (MIT) have shown that this technique has the potential to be built in to a biopsy needle, allowing doctors to instantaneously identify malignant or benign growths before an operation (23). Recent studies have indicated that the efficacy of these techniques can be enhanced by including a wider spectral region beyond 1800 cm-1 in the model to increase specificity (24). Finally, for the characterization of highly fluorescent biological tissues, such as liver and kidney samples, researchers have shown evidence to suggest that shifting the excitation laser wavelength to 1064 nm provides a cleaner Raman response with reduced fluorescence compared to excitation at 785 nm (25). Dispersive Raman technology at 1064 nm is relatively new and is slowly gaining traction. Most biological tissues produce fluorescence to a greater or lesser extent, so reducing this effect will improve the Raman signal and potentially improve the analysis and the quality or accuracy of the diagnostic outcome.
SERS Immunoassay for Pancreatic Cancer Biomarker Screening
Pancreatic adenocarcinoma is the fourth most common cause of cancer deaths in the United States, with a 5 year survival rate of ~6% and an average survival rate of <6 months. The reason for this high ranking is that the disease can only be detected and diagnosed by conventional radiological and histopathological detection methods when it has progressed to an advanced stage. After it has been diagnosed, resection (partial removal) of the pancreas is the normal treatment (26).
There are potentially more than 150 biomarkers found in serum for the detection of pancreatic and other cancers. One of the most prevalent is carbohydrate antigen 19-9 (CA 19-9), a mucin type glycoprotein sialosyl Lewis antigen (SLA), which shows elevated levels in ~75% of patients with pancreatic adenocarcinoma (27). The most common diagnostic test for these biomolecular compounds is enzyme-linked immunosorbent assay (ELISA), which is a well-established bioassay that uses a solid-phase enzyme immunoassay to detect the presence of an antigen in serum samples. It works by attaching the sample antigen to the surface of the sorbent. Then a specific monoclonal antibody, which is linked to the enzyme, is applied over the surface so it can bind to the antigen. In the final step, a substance containing the enzyme's substrate is added, which generates a reaction to produce a color change in the substrate (28). Even though this technique is widely used for bioassays, it does have some limitations with regard to the detection of cancer biomarkers. For example, early stage markers are present at levels well below current detection capabilities. In addition, 100–200 μL of sample is typically required to achieve a low enough limit of detection. Moreover, CA 19-9 is unlikely to be detected in patients with tumors smaller than 2 cm. It is further complicated by the fact that an estimated 15% of the population cannot even synthesize CA 19-9. The limitations in the ELISA approach (29) have led to the investigation of alternative methods, including an immunoassay based on SERS. This offers the exciting possibility of an alternative, faster way of detecting many different biomarkers in the same assay.
A portable Raman spectrometer (MiniRam, B&W Tek) was used in a recent study at the University of Utah's Nano Institute to detect and quantify the CA 19-9 antigen in human serum samples (30). It showed the advantages of generating a SERS-derived signal from a Raman reporter molecule attached to the biomolecule marker analyte using gold nanoparticles. This process is carried out using an analyte sandwich immunoassay in which monoclonal antibodies are covalently bound to a solid gold substrate to specifically capture the antigen from the sample. The captured antigen is in turn bound to the corresponding detection antibody. This complex is joined together with gold nanoparticles that are labeled with different reporter molecules, which serve as extrinsic Raman labels (ERLs) for each type of antibody. The presence of specific antigens is confirmed by detecting and measuring the characteristic SERS spectrum of the reporter molecule. Let's take a closer look at how this works in practice for these biomarkers and how the sample is prepared and read-out, in particular.
The first step is the preparation of the ERLs. This is done by coating a 60-nm gold nanoparticle with the Raman reporter molecule, which in this case is 5,5'-dithiobis(succinimidyl-2- nitrobenzoate (DSNB). This complex is then coated with a target or tracer antibody using N-hydroxysuccinimide (NHS) coupling chemistry. The DSNB serves two purposes: The nitrobenzoate group is the reporter molecule and the NHS group enables the protein to be coupled to the nanoparticle surface.
The second step is to prepare the substrate for capture. This is done by resistively evaporating solid gold under vacuum using thin film deposition on a glass microscope slide and then growing a monolayer of dithiobis(succinimidyl propionate) (DSP) on the surface of the slide to link the capture antibody to the gold surface. Next, the appropriate antibody is attached to the substrate, based on the biomarker of interest.
The third step is to make the immunoassay sandwich. This step involves incubating the slide with serum containing the CA 19-9 or MMP-7 antigens, which are then captured by surface-bound monoclonal antibodies and labeled with the ERLs.
The fourth and final step is to read out the Raman signal of the functional group of interest, using a spot size of ~85 μm with a laser excitation wavelength of 785 nm. In the study, the symmetric nitro stretch band of the DNSB molecule at 1336 cm-1 was used for quantitation by measuring the peak height to construct a calibration curve for various concentrations of the antigens spiked into serum. Real-world serum samples are then quantified using this calibration curve.
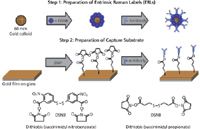
Figure 4: Preparation of labels and capture substrate. Adapted from reference 30.
The four steps of this procedure are shown in Figures 4 and 5, which show the preparation of the ERLs and capture substrate, together with the sandwich immunoassay and readout process. Using the nitro stretch band at 1336 cm-1, the SERS technique's detection capability for the CA 19-9 antigen was approximately 30–35 ng/mL, which was similar to that of the ELISA method. Preliminary results on real-world, human serum samples have shown that both techniques are generating very similar quantitative data, which is extremely encouraging if SERS is going to be used for the diagnostic testing of pancreatic and other types of cancers in a clinical testing environment. However, the investigation is still ongoing, particularly in looking for ways to lower the level of detection in human serum samples. Some of the procedures being investigated to optimize assay conditions include faster incubation times, different buffers, and the use of surfactants and blocking agents. Additionally, the bioassays in this study were initially carried out using an array format on a microscope slide, but have now been adapted to a standard 96-well plate format used for traditional clinical testing bioassays. By adopting these method enhancements, there is strong evidence that the SERS results could potentially be far superior and more cost effective than the ELISA method.
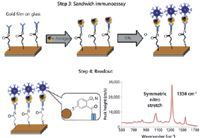
Figure 5: Sandwich immunoassay and readout process. Adapted from reference 30.
Conclusion
Both Raman spectroscopy and SERS are proving to be invaluable tools in the field of biomedical research and clinical diagnostics. The robust, compact, fit-for-purpose Raman spectrometer designs are appropriate for use in surgical procedures to help surgeons assess tumors and allow rapid decisions to be made. Raman systems are also being developed for molecular diagnostic testing to detect and measure human cancer biomarkers. Based on the SERS technique, this could potentially change the way bioassays are performed to improve both the sensitivity and reliability of testing. The two applications highlighted in this review together with other examples of the use of Raman spectrometry in biomedical research areas, such as the identification of bacterial infections, are clearly going to make the technique an important part of the medical toolbox, as we continually strive to improve diagnostic techniques and bring a better health care system to patients.
Dr. Katherine Bakeev is the director of analytical services and support at B&W Tek in Newark, Delaware. Michael Claybourn is a consultant with Biophotonics in Medicine in Chartres, France. Robert Chimenti is the marketing manager, at B&W Tek and an adjunct professor in the department of physics and astronomy at Rowan University in Glassboro, New Jersey. Robert Thomas is the principal consultant at Scientific Solutions Inc., in Gaithersburg, Maryland. Direct correspondence to: katherineb@bwtek.com
References
(1) E. Lozano Diaz and R.J. Thomas, Pharmaceutical Manufacturing (January 2013), http://www.pharmamanufacturing.com/articles/2013/006.html?page=1.
(2) B. Diehl, C.S. Chen, B. Grout, J. Hernandez, S. O'Neill, C. McSweeney, J.M. Alvarado, and M. Smith, Eur. Pharm. Rev., Non-destructive Materials Identification Supplement 17(5), 3–8 (2012).
(3) D. Yang and R.J. Thomas, Am. Pharm. Rev. (December 2012), http://www.americanpharmaceuticalreview.com/Featured-Articles/126738-The-Benefits-of-a-High-Performance-Handheld-Raman-Spectrometer-for-the-Rapid-Identification-of-Pharmaceutical-Raw-Materials/.
(4) M. Ribick and G. Dobler, Eur. Pharm. Rev., Non-destructive Materials Identification Supplement 17(5), 11–15 (2012).
(5) Vibrational Spectroscopy of Polymers: Principles and Practice, N.J. Everall, J. Chalmers, and P.R. Griffiths, Eds. (John Wiley and Sons, Chichester, United Kingdom, 2007).
(6) M.B. Fenn, P. Xanthopoulos, G. Pyrgiotakis, S.R. Grobmyer, P.M. Pardalos, and L.L. Hench, Advances in Optical Technologies Article ID 213783, Volume 2011.
(7) N. Jing, R.J. Lipert, G.B. Dawson, and M.D. Porter, Anal. Chem. 71(21), 4903–4908, (1999).
(8) Q. Tu and C. Chang, Nanomedicine 8, 545–558 (2012).
(9) A.A. Bhirde, G. Liu, A. Jin, R. Iglesias-Bartolome, A.A. Sousa, R.D. Leapman, J. Silvio Gutkind, S. Lee, and X. Chen, Theranostics 1, 310–321 (2011).
(10) L.-P. Choo-Smith, H.G.M. Edwards, H.P. Endtz, J.M. Kros, F. Heule, H. Barr, J.S. Robinson Jr., H.A. Bruining, and G.J. Puppels, Biopolymers 67(1), 1–9 (2002).
(11) Handbook of Raman Spectroscopy, I.R. Lewis and H.G.M. Edwards, Eds. (Marcel Dekker, New York, 2001).
(12) G. McNay, D. Eustace, W.E. Smith, K. Faulds, and D. Graham, Appl. Spectrosc. 65(8), 825–837 (2011).
(13) K. Kneipp and H. Kneipp, Appl. Spectrosc. 60, 322a (2006).
(14) R. Lewandowska, Raman Technology for Today's Spectroscopists supplement to Spectroscopy 28(6s), 32–42 (2013).
(15) E.C. Le Ru and P.G. Etchegoin, Principles of Surface-Enhanced Raman Spectroscopy and Related Plasmonic Effects (Elsevier B.V., Amsterdam, The Netherlands, 2009).
(16) Surface-Enhanced Raman Scattering – Physics and Applications 103, K. Kneipp, M. Moskovits, and H. Kneipp, Eds. (Springer, Berlin/Heidelberg, 2006).
(17) S. Botti, S. Almaviva, L. Cantarini, A. Palucci, A. Puiu, and A. Rufoloni, J. Raman Spectrosc. 44, 463–468 (2013).
(18) S-Y. Lin, M-J. Li, and W-T. Cheng, Spectroscopy 21(1), 1–30 (2007).
(19) J. Horsnell, P. Stonelake, J. Christie-Brown, G. Shetty, J. Hutchings, C. Kendalla, and N. Stone, Analyst 135, 3042–3047 (2010).
(20) C. Kallaway, L.M. Almond, H. Barr, J. Wood, J. Hutchings, C. Kendall, and N. Stone, Photodiagn. Photodyn. Ther. (2013), http://dx.doi.org/10.1016/j.pdpdt.2013.01.008.
(21) J.D. Horsnell, unpublished data (2010).
(22) J.D. Horsnell, J.A. Smith, M. Sattlecker, A. Sammon, J. Christie-Brown, C. Kendall,and N. Stone, The Surgeon 10(3), 123–127 (2011).
(23) A. Saha, I. Barman, N.C. Dingari, L.H. Galindo, A. Sattar, W. Liu, D. Plecha, N. Klein, R. Rao, R.R. Dasari, and M. Fitzmaurice, Anal. Chem. 84(15), 6715–6722 (2012).
(24) S. Duraipandian, W. Zheng, J. Ng, J.J. Low, A. Ilancheran, and Z. Huang, SPIE Proceedings Vol. 8572, Advanced Biomedical and Clinical Diagnostic Systems, March 2013.
(25) C.A. Lieber, H. Wu, and W. Yang, SPIE Proceedings Vol. 8572, Advanced Biomedical and Clinical Diagnostic Systems, March 2013.
(26) "Cancer Facts and Figures 2013," Atlanta, American Cancer Society, 2013.
(27) K.S. Goonetilleke and A.K. Siriwardena, Journal of Cancer Surgery 33, 266–270 (2007).
(28) Principles of ELISA: The ELISA Encyclopedia, http://www.elisa-antibody.com/ELISA-Introduction/.
(29) J.H. Granger, M.C. Granger, M.A. Firpo, S.J. Mulvihill, and M.D. Porter, The Analyst 138(2), 410–416 (2013).
(30) "Applications of Raman Spectroscopy in Biomedical Diagnostics," M. Kayat and J.H Granger, Spectroscopy on-line webinar, http://bwtek.com/webinar/applications-of-raman-spectroscopy-in-biomedical-diagnostics-spectroscopy/.

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
New AI Strategy for Mycotoxin Detection in Cereal Grains
April 21st 2025Researchers from Jiangsu University and Zhejiang University of Water Resources and Electric Power have developed a transfer learning approach that significantly enhances the accuracy and adaptability of NIR spectroscopy models for detecting mycotoxins in cereals.