Chemometrics in Spectroscopy Linearity in Calibration: Quantifying Non-linearity
This column presents results from some computer experiments designed to assess a method of quantifying the amount of non-linearity present in a dataset, assuming that the test for the presence of non-linearity already has been applied and found that a measurable, statistically significant degree of non-linearity exists.
In our last few columns (1–4), we discussed shortcomings of current methods used to assess the presence of non-linearity in data, and presented a new method that addresses those shortcomings. This new method is statistically sound, provides an objective means to determine if non-linearity is present in the relationship between two sets of data, and is inherently suitable for implementation as a computer program.

Jerome Workman Jr. & Howard Mark
A shortcoming of the method presented is one that it has in common with virtually all statistical tests: while it provides a means of unambiguously and objectively determining the presence of non-linearity, if we find that non-linearity is present, it does not address the question of how much non-linearity is present. This column, therefore, presents results from some computer experiments designed to assess a method of quantifying the amount of non-linearity present in a data set, assuming that the test for the presence of non-linearity already has been applied and found that indeed, a measurable, statistically significant degree of non-linearity exists.
The spectroscopic community, and indeed, the chemical community at large, is not the only group of scientists concerned with these issues. Other scientific disciplines also are concerned with ways to evaluate methods of chemical analysis. Notable among them are the pharmaceutical communities and the clinical chemistry communities. In those communities, considerations of the sort we are addressing are even more important, for at least two reasons:
- These disciplines are regulated by governmental agencies, especially the Food and Drug Administration. In fact, it was considerations of the requirements of a regulatory agency that created the impetus for this series of columns in the first place (1).
- The second reason is what drives the whole effort of ensuring that everything that is done, is done "right." An error in an analytical result can conceivably, in literal fact, cause illness or even death.
Thus, the clinical chemistry community also has investigated issues such as the linearity of the relationship between test results and actual chemical composition, and an interesting article provides the impetus for creating a method of assessing the degree of non-linearity present in the relationship between two sets of data (5).
Degree of Non-linearity
The basis for this calculation of the amount of non-linearity is illustrated in Figure 1. In Figure 1a, we see a set of data showing some non-linearity between the test results and the actual values. If a straight line and a quadratic polynomial both are fit to the data, then the difference between the predicted values from the two curves gives a measure of the amount of non-linearity. Figure 1a shows data subject to both random error and nonlinearity, and the different ways linear and quadratic polynomials fit the data. As shown in Figure 1a, at any given point, there is a difference between the two functions that represents the difference between the Y-values corresponding to a given X-value.
Figure 1b shows that irrespective of the random error of the data, the difference between the two functions depends only upon the nature of the functions and can be calculated from the difference between the Y-values corresponding to each X-value. If there is no non-linearity at all, then the two functions will coincide, and all the differences will be zero. Increasing amounts of non-linearity will cause increasingly large differences between the values of the two functions corresponding to each X-value, and these can be used to calculate the nonlinearity.
The calculation used is the calculation of the sum of squares of the differences (5). This calculation normally is applied to situations where random variations are affecting the data, and indeed, is the basis for many of the statistical tests that are applied to random data. However, the formalism of partitioning the sums of squares, which we have discussed previously (6) (also in [7], page 81 in the first edition or page 83 in the second edition) can be applied to data in which the variations are due to systematic effects rather than random effects. The difference is that the usual statistical tests (t, χ2, F, etc.) do not apply to variations from systematic causes because they do not follow the required statistical distributions. Therefore, it is legitimate to perform the calculation, as long as we are careful how we interpret the results.
Performing the calculations on function fitted to the raw data has another ramification: the differences, and therefore the sums of squares, will depend upon the units in which the Y-values are expressed. It is preferable that functions with similar appearances give the same computed value of non-linearity regardless of the scale. Therefore, the sum-of-squares of the differences between the linear and quadratic functions fitted to the data is divided by the sum-of-squares of the Y-values that fall on the straight line fitted to the data. This cancels the units, and therefore, the dependency of the calculation upon the scale.
A further consideration is that the value of the calculated non-linearity will depend not only upon the function that fits the data, but we suspect that it also will depend upon the distribution of the data along the X-axis. Therefore, for pedagogical purposes, here we will consider the situation for two common data distributions: the uniform distribution and the normal (Gaussian) distribution.
Figure 2 presents some quadratic curves containing various amounts of non-linearity. These curves represent data that was, of course, created synthetically. The purpose of generating these curves was for us to be able to compare the visual appearance of curves containing known amounts of non-linearity with the numerical values for the various test parameters that describe the curves. Figure 2 represents data having a uniform distribution of X-values, although, of course, data with a different distribution of X-values would follow the same curves. The curves were generated as follows: 101 values of a uniformly distributed variable (used as the X-variable) was generated by creating a set of numbers from 0 to 1 at steps of 0.01. The Y-values for each curve were generated by calculating the Y-value from the corresponding X-value according to the following formula:
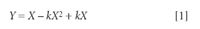
The parameter k in Equation 1 induces a varying amount of non-linearity in the curve. For the curves in Figure 1, k varied from 0 to 2 in steps of 0.2. The subtraction of the quadratic term in Equation 1 gives the curves their characteristic of being convex upward, while adding the term kX back in ensures that all the curves, and the straight line, meet at zero and at unity.
Table I presents the results of computing the linearity evaluation results for the curves shown in Figure 1, for the case of a uniform distribution of data along the X-axis. It presents the coefficients of the linear models (straight lines) fitted to the several curves of Figure 1, the coefficients of the quadratic model, the sum-of-squares of the differences between the fitted points from the two models, and the ratio of the sum-of-squares of the differences to the sum-of-squares of the X-data itself, which, as stated earlier, is the measure of non-linearity. Table I also shows the value of the correlation coefficient between the linear fit and the quadratic fit to the data, and the square of the correlation coefficient.

Table I. Uniform Data Distribution
In Table I, we see an interesting result — the ratio of sums of squares we are using for the linearity measure is equal to 1 (unity) minus the square of the computed correlation coefficient value between the linear fit and quadratic fit to the data. This should not surprise us. As noted previously, the same formalisms that apply to random data also can be applied to data in which the differences are systematic. Therefore, the equality we see here corresponds to the well-known property of sums of squares from any regression analysis, that from the analysis of variance of the regression results, the correlation coefficient is related to sum-squared-error of the analysis in a similar way (8). It also is interesting to note that the coefficients of the models resulting from the calculations on the data (shown in Figure 1) are not the same as the original generating functions for the data. This is because the generating functions (from Equation 1) are not the best-fitting functions (nor, as we shall see, are they the orthogonalized functions), which is what is used to create the models, and the predicted values from the models.

Figure 1. An illustration of the method of measuring the amount of non-linearity. (a) Hypothetical synthetic data to which each of the functions are fit. (b) The functions, without the data, show the differences between the functions at two values of X. The circles show the value of the straight line; the crosses show the value of the quadratic function at the given values of X.
A New Calculation
Because the correlation coefficient is an already-existing and known statistical function, why is there a need to create a new calculation for the purpose of assessing non-linearity? First, the correlation coefficient's roots in statistics directs the mind to the random aspects of the data for which it normally is used. In contrast, therefore, using the ratio of the sum of squares helps to remind us that we are dealing with a systematic effect whose magnitude we are trying to measure, rather than a random effect for which we want to ascertain statistical significance.
Secondly, as a measure of non-linearity, the calculation conforms more closely to that concept than does the correlation coefficient. As a contrast, we can consider terms such as precision and accuracy, where "high precision" and "high accuracy" mean data with small values of <whatever measure is used, such as standard deviation> while "low precision" and "low accuracy" mean large values of the measure. Thus, for those two characteristics, the measured value changes in opposition to the concept. If we were to use the correlation coefficient calculation as the measure of non-linearity, we would have the same situation. However, by defining the "linearity" calculation the way we did, the calculation now runs parallel to the concept — a calculated value of zero means "no non-linearity," while increasing values of the calculation correspond to increasing non-linearity.
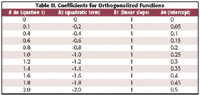
Table II. Coefficients for Orthogonalized Functions
Another interesting comparison is between the coefficients for the functions representing the best-fitting models for the data, and the coefficients for the functions that result from performing the linearity test as described in the previous column (4). We have not looked at these before because they are not involved directly in the linearity test. Now, however, we consider them for their pedagogic interest. These coefficients, for the case of testing a quadratic non-linearity of the data from Figure 1, are listed in Table II. We note that the coefficients for the quadratic terms are the same in both cases. However, the best-fitting functions have a constant intercept and varying slopes, while the functions based upon the orthogonalized quadratic term have a constant slope and varying intercept.

Figure 2. Curves illustrate varying amounts of non-linearity.
We now take a look at the linearity values obtained when the X-data is normally distributed. The non-linearity used is the same as that used earlier in the case of uniformly distributed data, and the same diagram (Figure 2) applies, so we need not reproduce it. The difference is that the X-data is normally distributed, so that there are more samples at X = 0.5 than at the extremes of the range of Figure 2, the falloff varying appropriately. The standard deviation of the X -values used was 0.2, so that the ends of the range corresponded to ±2.5 standard deviations. Again, synthetic data at the same 101 values of X were generated. In this case, however, multiple data at each X-value were created, the number of data at each X-value being proportional to the value of the normal distribution corresponding to that X-value. The total number of data points generated, therefore, was 5468.
We can compare the values in Table III with those in Table I; the coefficients of the models are almost the same. The coefficients for the quadratic model are, unsurprisingly, identical in all cases, because the data values are identical and there is no random error. The main difference in the linear model is the value of the intercept, reflecting the higher average value of the Y-data resulting from the center of the curves being weighted more heavily. The sums-of-squares are necessarily larger, simply because there are more data points contributing to this sum.
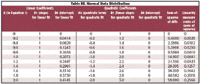
Table III. Normal Data Distribution
The interesting (and important) difference is in the values for the ratio of sums-of-squares, which is the non-linearity measure. As we see, at small values of non-linearity (that is, k = 0, 1, 2), the values for the non-linearity values are almost the same. As k increases, however, the value of the non-linearity measure decreases for the case of normally distributed data, as compared to the uniformly distributed data, and the discrepancy between the two gets greater as k continues to increase. In retrospect, this also should not be surprising, because in the normally distributed case, more data is near the center of the plot, and therefore, in a region where the local non-linearity is smaller than the non-linearity over the full range. Therefore, the normally distributed data is less subject to the effects of the non-linearity at the wings, because less of the data is there.
As a quantification of the amount of non-linearity, we see that when we compare the values of the non-linearity measure between Tables I and III, they differ. This indicates that the test is sensitive to the distribution of the data. Furthermore, the disparity increases as the amount of curvature increases. Thus, this test, as it stands, is not completely satisfactory because the test value does not solely depend upon the amount of non-linearity, but also upon the data distribution.
In our next column, we will consider a modification of the test that will address this issue.
Jerome Workman Jr. serves on the Editorial Advisory Board of Spectroscopy and is director of research, technology, and applications development for the Molecular Spectroscopy & Microanalysis division of Thermo Electron Corp. He can be reached by e-mail at: jerry.workman@thermo.com Howard Mark serves on the Editorial Advisory Board of Spectroscopy and runs a consulting service, Mark Electronics (Suffern, NY). He can be reached via e-mail at: hlmark@prodigy.net
References
1. H. Mark and J. Workman, Spectroscopy 20(1), 56–59 (2005).
2. H. Mark and J. Workman, Spectroscopy 20(3), 34–39 (2005).
3. H. Mark and J. Workman, Spectroscopy 20(4), 38–39 (2005).
4. H. Mark and J. Workman, Spectroscopy 20(9), 26–35 (2005).
5. M.H. Kroll and K. Emancipator, Clinical Chemistry 39(3), 405–413 (1993).
6. J. Workman and H. Mark, Spectroscopy 3(3), 40–42 (1988).
7. H. Mark and J. Workman, Statistics in Spectroscopy, 1st ed. (Academic Press, New York, 1991).
8. C. Daniel and F. Wood, Fitting Equations to Data — Computer Analysis of Multifactor Data for Scientists and Engineers, 1st ed. (John Wiley & Sons, Hoboken, NJ, 1971).

LIBS Illuminates the Hidden Health Risks of Indoor Welding and Soldering
April 23rd 2025A new dual-spectroscopy approach reveals real-time pollution threats in indoor workspaces. Chinese researchers have pioneered the use of laser-induced breakdown spectroscopy (LIBS) and aerosol mass spectrometry to uncover and monitor harmful heavy metal and dust emissions from soldering and welding in real-time. These complementary tools offer a fast, accurate means to evaluate air quality threats in industrial and indoor environments—where people spend most of their time.
Smarter Sensors, Cleaner Earth Using AI and IoT for Pollution Monitoring
April 22nd 2025A global research team has detailed how smart sensors, artificial intelligence (AI), machine learning, and Internet of Things (IoT) technologies are transforming the detection and management of environmental pollutants. Their comprehensive review highlights how spectroscopy and sensor networks are now key tools in real-time pollution tracking.
New AI Strategy for Mycotoxin Detection in Cereal Grains
April 21st 2025Researchers from Jiangsu University and Zhejiang University of Water Resources and Electric Power have developed a transfer learning approach that significantly enhances the accuracy and adaptability of NIR spectroscopy models for detecting mycotoxins in cereals.