Classical Least Squares, Part IV: Spectroscopic Theory Continued
The connection between the mathematics of classical least squares and the graphical displays used to present it is examined in further detail.

This column is a continuation of our discussion of the classical least squares (CLS) approach to calibration (1–3). As we usually do when we continue the discussion of a topic through more than one column, we continue the numbering of equations from where we left off in the last installment.
The insight we are trying to develop hinges on Figure 1 (which first appeared as Figure 5 in Part III of this series [3]) and the meaning of it in terms of equating the concepts of the spectroscopic and mathematical views of Beer's law as it applies to spectra measured for the purpose of calibrations for quantitative analysis. Therefore, we also repeat equation 24:
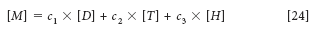
where [M], [D], [T], and [H] represent the spectra (which are now vectors, in this representation) of the mixture, dichloromethane, toluene, and n-heptane, respectively, and the corresponding c1, c2, and c3 represent the concentrations of dichloromethane, toluene, and n-heptane, respectively.
The reason for all this is the final sentence in our previous column (3): "Figure 5 is where the spectroscopy meets the math." We also repeat some of the prior explanation: In this figure we have taken Figure 4 from Part III of this column series and added some symbols to it. We now compare the figure (Figure 1 in this installment) to equation 24. In equation 24, we represented the spectra of each of the components of the mixture by a symbol ([D], [T], [H]), each representing the corresponding spectrum.

Figure 1: Absorbance spectra of pure dichloromethane, n-heptane, and toluene, and of a ternary mixture of the three (x-axis in wavenumbers), along with the concentration information that makes this the graphical representation of equation 24.
In Figure 1 we have effectively rewritten equation 24 by replacing the symbol representing each spectrum by the actual spectrum.
That's why Figure 1 is where the spectroscopy meets the math. Figure 1 is the same as equation 24, except that the spectra, which are indicated by the matrix symbols [D], [T], and [H] in equation 24, are shown in their conventional graphical form in Figure 1.
This can be even further emphasized by replacing the graphical presentation of the spectra with the actual numbers they represent, as we just described them. In Table I we present the numbers that make up the four spectra that concern us here: the spectra of the mixture of toluene, dichoromethane, and n-heptane, and the absorbances of the three pure materials, whose spectra are presented in Figure 1.
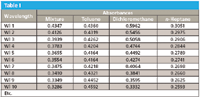
Table I
Now we can go one step further, and add to Table I the expressions needed to convert the bare listing of values into a set of equations describing the behavior of the data. We do so in Table II.
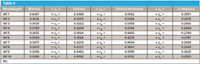
Table II
Note that the heading of Table II is essentially a rewriting of equation 23 (3), and each row of Table II represents the calculations that are to be done for each wavelength. With Table II, therefore, we have come full circle. Were we to rewrite Table II in matrix notation, we would arrive back at equation 24, which, as we showed immediately before, was the matrix equation corresponding to the computations to be done.
An alternate view of the sequence we went through, which we want to emphasize here, is that we arrived at Table II, and thereby at equation 24, by starting with the graphical presentation of the spectra of some pure components and a mixture made up from those pure components (Figure 1 from reference 1), and from that we noted that the spectra are in fact made up of the numbers representing those spectra at each wavelength.
So we've found that the absorbances of the various materials, when added together wavelength by wavelength, give the absorbances of the mixture. This ties together (in a mathematical sense) the absorbances of the three different materials. Is there anything else that ties together the absorbances of each material, in a similar sense?
Yes, there is. There is the fact, shown in Figure 1 and in Table II, that all the absorbances for toluene are multiplied by the same factor (the weighting factor, representing the concentration of toluene) indicating its contribution to the mixture spectrum. Similarly, all the absorbances of dichloromethane are multiplied by the same factor (representing the concentration of dichloromethane), as are all the absorbances of n-heptane multiplied by the same factor (representing the concentration of n-heptane.
So now we know how to synthesize the spectrum of a mixture of various materials: We add together the (weighted) spectra of the materials making up the mixture, just as specified in equation 23.
This is all of some mild interest, intellectually and pedagogically, but it's not what we want to do. What we really want to do is the inverse operation: to take the spectrum of a mixture, and from it determine the concentrations of the various components of the spectrum. How can we do that?
To start getting an appreciation for how we will do that, we again look at equations 23 and 24. Equation 23 tells us that the concentrations are the weighting factors, such that when multiplied by the absorbances, and then added together, the sum is the absorbance of the mixture. Equation 24 tells us that this is true for the absorbances at all the wavelengths; furthermore, the absorbances are linked together by two links. The first link is physical: The absorbances are representative of the same specific compounds that give rise to them and are represented by the same concentration of each of the materials. The second link is mathematical: All the absorbances are multiplied by the same coefficient of the equation representing the spectroscopic system.
So to give us a head-start, let's ask the following question: Where else do we have a situation where a number of variables are each multiplied by a coefficient — and the same coefficient is used to weight the same variable in each sample — to produce a numeric result?
The answer is when we do multiple linear regression (MLR) (or inverse least squares [ILS]) calibration. There's an exact parallelism between CLS and ILS. This parallelism is made explicit in Table III.

Table III
In Table III, we have taken the same set of numbers and inserted them twice in the table, once in the context of CLS and once in the context of ILS, changing only the column headings, which changes the meanings of the numbers. We list the comparisons in Table IV.
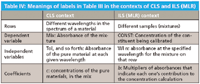
Table IV: Meanings of labels in Table III in the contexts of CLS and ILS (MLR)
So we see that, first of all, the same quantities are present in each context: absorbances of mixtures, concentrations, coefficients. The only quantity missing is that in the ILS context, there are no absorbances of pure materials explicitly provided.
So what does this all mean in terms of how we want to use this information? First let's look at the ILS section of Table III. When we do an MLR or ILS calibration, we regress all the independent variables against the dependent variable; that's naturally what a regression is all about. So what we would want to do is to take the information representing the independent variables in the CLS case and relate them to the independent variable just as we did in the ILS case. In the ILS case we used least-squares mathematics to make the comparison that created the relationships, and we want to do the same type of least-squares computations here to relate the information in the independent variables in the CLS case to the dependent variable in that case. Therefore we would do exactly the same calculations using the independent variables in the CLS case that we did in the ILS case.
The difference is in what the variables in the CLS case represent. In the CLS case, the dependent variable represents the spectrum of the mixture. The independent variables are the spectra of the materials that went into the mixture. Therefore what we do is to regress the spectra of the pure materials against the spectrum of the mixture. This will tell us what the coefficient is that represents the amount of each material that appears in the mixture. This coefficient, then, is the concentration of that material in the mixture.
The concentration of the materials in the mixture is exactly the information that we want to get out of the analysis. Now, as we have seen, this concentration is exactly the result of performing the least-squares calculations using the spectra of the pure materials against the spectrum of the mixture.
Thus we see that the information we want falls naturally out of the calibration calculations when we perform them the CLS way. We also note that coefficients are exactly the same coefficients that were used in equations 21 and 23 to create the spectrum of the mixture out of the spectra of the components to start with. We see, therefore, that doing the least squares calculations is the way to recover the concentrations of the materials whose spectra comprise the spectrum of the mixture.
This is the explanation for the similarity of the expressions in the two entries of Table I from reference 3. They both represent least squares calculations. The differences between the two expressions arise from the difference in the different locations of those different variables that are used in the calculations in the two cases. These reflect the differences in the meaning of the different variables that are used in the two calculations.
In the case of ILS, the concentration of the analyte must be known beforehand. The coefficients that are calculated represent the contribution of the analyte to the spectrum plus corrections for contributions to the spectrum made by the other materials in the sample.
In the case of CLS, no concentration information need be known beforehand. The concentration information is calculated ab initio, purely from the spectral information available. The other side of that coin, however, is that instead of concentration information from the sample, we need to know the spectra of the pure materials that comprise the sample mixture.
The more alert among our readers might ask a question: Why is it that if Table I from reference 3 specifies the same calculations for both cases, the matrix operations are written almost in reverse order from one another? The answer, though it might seem a priori puzzling, turns out to be actually rather simple and mundane, indeed almost trivial.
Recall that when we rearranged the data to form Figures 4 and 5 in reference 3 (and Figure 1 in this installment), we rotated the graphical presentation of the spectra by 90° (compare Figure 2b to Figure 5 in reference 3). In terms of the actual numerical data, presented as a matrix, this rotation transformed the rows of the matrix into columns and vice versa (that is, we performed a transpose operation on the matrix). This requires that, if we wish to perform similar computations on the data before and after the transposition, the order of multiplication must be reversed to compensate. Does this give us the same result for the new computation? Well, almost. Having both transposed the matrix and reversed the order of computations, we will wind up with the transpose of the original result. This is a theorem of matrix math that we don't usually have much need for, but because it has affected our discussion, we felt a need to explain it.
Howard Mark serves on the Editorial Advisory Board of Spectroscopy and runs a consulting service, Mark Electronics (Suffern, NY). He can be reached via e-mail: hlmark@prodigy.net

Howard Mark
Jerome Workman, Jr. serves on the Editorial Advisory Board of Spectroscopy and is currently working in the medical device industry using spectroscopy. His email address is: JWorkman04@gsb.columbia.edu

Jerome Workman, Jr.
References
(1) H. Mark and J. Workman, Spectroscopy 25(5), 16–21 (2010).
(2) H. Mark and J. Workman, Spectroscopy 25(6), 20–25 (2010).
(3) H. Mark and J. Workman, Spectroscopy 25(10), 22–31 (2010).

AI Shakes Up Spectroscopy as New Tools Reveal the Secret Life of Molecules
April 14th 2025A leading-edge review led by researchers at Oak Ridge National Laboratory and MIT explores how artificial intelligence is revolutionizing the study of molecular vibrations and phonon dynamics. From infrared and Raman spectroscopy to neutron and X-ray scattering, AI is transforming how scientists interpret vibrational spectra and predict material behaviors.
Real-Time Battery Health Tracking Using Fiber-Optic Sensors
April 9th 2025A new study by researchers from Palo Alto Research Center (PARC, a Xerox Company) and LG Chem Power presents a novel method for real-time battery monitoring using embedded fiber-optic sensors. This approach enhances state-of-charge (SOC) and state-of-health (SOH) estimations, potentially improving the efficiency and lifespan of lithium-ion batteries in electric vehicles (xEVs).
New Study Provides Insights into Chiral Smectic Phases
March 31st 2025Researchers from the Institute of Nuclear Physics Polish Academy of Sciences have unveiled new insights into the molecular arrangement of the 7HH6 compound’s smectic phases using X-ray diffraction (XRD) and infrared (IR) spectroscopy.