Posttranslational Modification Characterization via Electron Capture Dissociation Using a Linear Ion Trap Time-of-Flight Mass Spectrometer
Special Issues
The authors introduce a compact ECD device coupled to a linear ion trap time-of-flight instrument, and use it to analyze protein phosphorylation in both offline and online modes.
Successful characterization of protein posttranslational modifications (PTMs) is critical to our understanding of many biological processes. Unfortunately, attempts to describe PTMs often prove experimentally difficult and result in ambiguous conclusions. As technologies in the field of mass spectrometry (MS) continue to improve, people are turning increasingly to mass spectral techniques for PTM characterization. Recently, novel modes of peptide fragmentation have emerged that are giving scientists greater ability to elucidate protein posttranslational modification. One example is electron-capture dissociation (ECD), an alternative fragmentation mechanism for use in peptide analysis via tandem mass spectrometry. ECD selectively cleaves N-C(alpha) bonds of the peptide backbone, yielding c- and z-ions without the loss of labile PTMs. ECD therefore holds advantages over conventional fragmentation techniques such as collisionally induced dissociation (CID), which often cleave PTMs from the peptide backbone, precluding the ability to characterize them. Until recently, however, ECD was limited to resource-intensive Fourier transform–ion cyclotron resonance instruments. Here, we introduce a compact ECD device coupled to a linear ion trap time-of-flight instrument, and use it to analyze protein phoshorylation in both offline (via direct infusion) and online (via nanoLC infusion) modes. We also utilize ECD to characterize both N- and O-linked peptide glycosylations. The results demonstrate ECD's capability to analyze a variety of posttranslational modifications using linear ion trap TOF-MS.
The majority of proteins are modified covalently in some manner prior to carrying out their role in the body. The nature of these modifications can vary significantly in both size and complexity, ranging from simple methylation and phosphorylation through more complex glycosylation and ultimately to small peptides and proteins such as ubiquitination and SUMOylation. Post-translational modifications (PTMs) are involved in a wide range of fundamental cellular processes, including enzyme regulation, signal cascade control, and cell–cell recognition and adhesion. PTMs are thereby likely involved in the mechanism of many disease classes, and are of great interest to the biochemical community.
Traditionally, tandem mass spectrometry (MS-MS) has utilized collisionally induced dissociation (CID) to provide information on both protein sequence and post-translational modifications. CID-MS-MS produces b- and y-ions by activating the precursor ion collisionally with inert gas molecules. Unfortunately, information on the labile PTM often is lost during the CID process. For example, phosphorylated serine and threonine residues, which frequently are involved in a variety of intracellular signaling processes, have a tendency to lose their modifier phosphate groups via the β-elimination of a phosphoric acid molecule.
In 1998, Zubarev and colleagues (1) reported electron-capture dissociation (ECD) as a new dissociation technique. ECD induces cleavage of peptide backbone bonds of multiply charged peptides and proteins via low energy (<1 eV) electron capture. While CID is induced by the excitation of vibrational energy in the analyte ions, ECD proceeds via intramolecular electron transfer (Figure 1). ECD randomly cleaves N-Cα bonds of the peptide backbone and yields c- and z-ions, regardless of their sequence, without the loss of labile modifications. ECD therefore allows both peptide identification and precise determination of the PTM location.
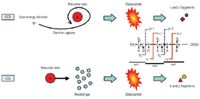
Figure 1: A schematic illustration of ECD versus CID MS-MS. ECD-MS-MS cleaves the N-Cα bonds, and generates "c" and "z" fragment ions, while leaving the labile PTM intact.
Although ECD has been recognized as a powerful tool for PTM analysis, traditionally it has required a Fourier-transform ion cyclotron resonance (FT-ICR) mass spectrometer equipped with an electrostatic magnetic field to provide low energy electrons. Such instruments are resource-intensive, require expert skill for operation, and are large and expensive. Moreover, ECD using FT-ICR MS requires long acquisition times because the intensities of the fragment ions are weak. For these reasons, the use of ECD has been limited in the field of basic research.
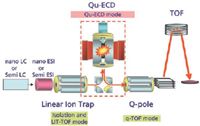
Figure 2: A schematic diagram of the linear ion trap instrument equipped with a QuECD device between the linear ion trap, which isolates precursor ions and performs CID, and the quadrupole collision-induced dissociation (QCID) cell.
In 2004, Baba and colleagues (2) succeeded in the achievement of ECD in a radio-frequency (RF) ion trap by using a linear ion trap device with a cylindrical permanent magnet. This small device, named QuECD, can be coupled to a linear ion trap TOF instrument to create a platform capable of high-throughput ECD (Figure 2) (2,3). Figure 3 shows the detailed structure of the ECD device. To avoid electron heating by the RF electric field in the ion trap, the electron beam is directed along the central axis of the linear ion trap. A magnetic field parallel to the central axis promotes electron travel along the central axis. The speed of the QuECD reaction is faster than conventional ECD using FT-ICR MS and is roughly equal to conventional CID. Because the reaction occurs on the chromatographic timescale, it can be performed during a standard liquid chromatography (LC)–MS experiment, which was unachievable previously with FT-ICR-based ECD techniques.
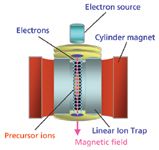
Figure 3: A schematic diagram of the QuECD device. Low-energy electrons introduced from the top of the device are accelerated by a permanent magnet surrounding a radio-frequency linear ion trap.
Here we use QuECD coupled with a linear ion trap TOF instrument (NanoFrontier eLD, Hitachi High Technologies America, Pleasanton, California) to analyze protein phosphorylation in both direct infusion and nanoLC modes. We also utilize this platform to characterize both N- and O-linked peptide glycosylations. The results demonstrate the capability of QuECD, in conjunction with linear ion trap TOF MS, to analyze a variety of PTMs.
Characterization of Phosphopeptides
Direct Infusion
Conventional CID of phosphorylated peptides induces neutral loss of phosphoric acid, complicating determination of the peptide sequence and the location of phosphorylation. The analysis becomes increasingly difficult for multiply phosphorylated peptides. Figure 4 shows QuECD spectra of mono- and diphosphorylated versions of a peptide containing three tyrosine residues. The spectra indicate that Tyr12 of the monophosphorylated peptide and Tyr11 and Tyr12 of the diphosphorylated peptide are modified. These data demonstrate the utility of QuECD for determining the PTM locations of singly and multiply modified peptides.

Figure 4: QuECD spectra of (a) mono- and (b) diphosphorylated peptides. Tyr12 in (a) and Tyr11 and Tyr12 in (b) are phosphorylated, respectively.
NanoLC Infusion
As mentioned previously, the QuECD reaction occurs on the same timescale as conventional CID, which enables ECD to be performed during a standard LC–MS experiment. We used both LC–CID-MS–MS and LC–ECD-MS-MS to examine the phosphoproteome of an HEK293T cell lysate. Figure 5 compares sample MS-MS spectra of a triply charged phosphopeptide at m/z 612.3 observed by both CID and ECD. In the case of CID-MS-MS, the loss of the phosphate group has impeded further fragmentation of the peptide. The resulting inadequate fragmentation pattern observed in the CID-MS-MS spectrum is in sharp contrast to the more complete fragmentation pattern observed via ECD-MS-MS. Using the ECD-MS-MS spectrum, this ion was identified as a peptide from an insulin-like growth factor II receptor that contains a phosphorylation at Serine 2484.
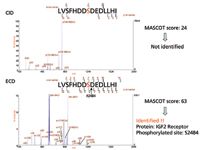
Figure 5: Comparison of typical CID- and QuECD-MS-MS spectra encountered in the phosphoproteome study of an HEK293T cell lysate. Both spectra are product ion spectra of the precursor ion [M + 3H]3+ at m/z 612.3 that correspond to a peptide spanning residues 2477 to 2491 of the human protein IGF2R. The newly identified phosphoserine at position 2484 is denoted in red type.
We found that the combination of CID and ECD data together provides the most comprehensive analysis of phosphorylated peptides. The peak lists obtained by CID- and ECD-MS-MS experiments were merged, and the resulting composite data set was searched against the Swiss-Prot database using the MASCOT search engine. More phosphorylated proteins and peptides were identified using the combined data set than those from individual searches. These results demonstrate that ECD is an effective technique complementary to CID and that a combination of the two dissociation modes provides an excellent platform for phosphoproteome analysis.
Characterization of Glycopeptides
To identify a glycopeptide, the following three factors must be determined: the amino acid sequence of the peptide, the location of glycosylation, and the carbohydrate structure. ECD simultaneously identifies an amino acid sequence and glycosylation sites by fragmenting peptides while leaving the carbohydrate chain intact. The ECD spectrum in Figure 6a shows data generated from an N-linked glycopeptide. It suggests that the interval between C3 and C4 ions exactly corresponds to the sum of the molecular weight of N-glycan coupled with asparagine. The amino acid sequence and binding site (N) are both determined without dissociating the carbohydrate chain. Figure 6b illustrates the analysis of an O-linked glycopeptide with three possible glycosylation sites (S). The ECD spectra suggest that the interval between z7 and z6 (or between c5 and c6) ions exactly corresponds to the sum of the molecular weights of the O-glycan coupled with a serine, uniquely determining the glycan binding site. Again, the amino acid sequence is clearly determined from c- and z-type fragment ions without dissociating the carbohydrate chain.

Figure 6: QuECD spectra of (a) N-linked and (b) O-linked glycopeptides.
Conclusion
The results indicate that QuECD, coupled with the linear ion trap TOF MS, creates an efficient tool for analysis of PTMs such as phosphorylation and glycosylation. The results also show that the combination of CID and ECD data result in improved reliability and depth of phosphoproteome analysis. Because the QuECD reaction occurs on the chromatographic timescale, this platform is capable of running LC–ECD-MS-MS experiments, allowing for ECD interrogation of complex samples.
Acknowledgment
The authors would like to thank Naomi Manri of the Central Research Laboratory, Hitachi Ltd., for his help.
M. Alexander Shaw, Akira Tsuboyama, and Chad Ostrander are with Hitachi High-Technologies America, Pleasanton, California.
Masaki Watanabe is with Hitachi High-Technologies, Hitachinaka, Ibaraki, 312-8504, Japan.
Takuji Nabetani and Yoshio Hirabayashi are with Brain Science Institute, RIKEN, Wako, Saitama, 351-0198, Japan.
References
(1) R.A. Zubarev et al., J. Am. Chem. Soc. 120(13), 3265–3266 (1998).
(2) T. Baba et al., Anal. Chem. 76(15), 4263–4266 (2004).
(3) H. Satake et al., Anal. Chem. 79(22), 8755–8761 (2007).

Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.