Advances in TOF-MS-Based Screening for Food Safety Residue Analysis with a Positive Approach
Special Issues
Mass spectrometry plays an increasingly significant role in the analysis of residues and contaminants in food. Here we will illustrate how the combination of ultrahigh-pressure liquid chromatography (UHPLC) and high-resolution time-of-flight-mass spectrometry (TOF-MS) is used to generate a screen of veterinary drug residues in products of animal origin. The use of UHPLC–TOF-MS and dedicated, workflow directed software allows rapid screening for large numbers of residues and automated quantification of positive samples. In addition, we illustrate how the data generated using MSE acquisition mode enable critical structural information to be collected, which offers additional selectivity and confirmatory data for compound identification and facilitates elucidation of the structure of newly discovered compounds.
Food safety is a key concern of governments, food producers, and manufacturers. Recent events involving compounds such as melamine (1) and dioxins (2) highlight the risks posed by poor agricultural practices, accidental contamination, and deliberate adulteration. Heightened vigilance goes hand-in-hand with more effective and efficient solutions for the comprehensive screening of food products for chemicals that might pose a risk and to improve the detection and identification of new or unexpected compounds of concern.
Mass spectrometry (MS) is playing a significant role in residue testing. Tandem quadrupole MS-MS has become the accepted, and in some regulatory frameworks, the required technique, for quantitative analysis and confirmation of identity. The main limitation of tandem quadrupole MS-MS and of the targeted nature of so many of the experimental methods of analysis is the number of residues that can be included in a single experiment due to the nature of quadrupole-based instrument duty cycles. Targeting specific residues in an MS-MS experiment on a tandem quadrupole instrument does provide high selectivity and enables accurate quantification of compounds at trace levels even in complex food matrices. However, the nature of the experiment means that data is collected only for those compounds that have been targeted. There is no capability to look at and detect untargeted compounds, meaning potential contaminants can escape detection. Also, data cannot be reinterrogated historically postacquisition for the presence of additional compounds that might be relevant. This is one reason that analysts seek to target the maximum number of compounds with multiresidue methods that can target hundreds of compounds (3).
There is now a clear requirement for very large multiresidue screens and the desire to capture "all of the data, all of the time" thereby generating as comprehensive a dataset as possible. This has become a major driver in the application of high-resolution MS, notably time of flight (TOF)-MS, to food safety residue and contaminant screening and discovery of new or emerging compounds of concern. Here we present a workflow for a generic TOF-MS screening scenario and illustrate it with an example of the targeted screening of animal products for veterinary drug residues.
The workflow shown in Figure 1 describes a generic TOF-MS screening scenario, in which a nonselective sample preparation is used to generate an extract suitable for screening analysis. Such a nonselective extraction can maximize the number of residues that can be determined but results in a complex sample extract with many coextracted matrix components. This places extreme demands upon the ability to separate and detect each component. The quality of the final result will be directly related to the quality of the initial chromatographic separation. The better that matrix and residue components are separated from one another, the more effective detection is and the better the data obtained. The introduction of columns packed with sub-2-μm particles in 2004, and the development of a new generation of instruments capable of maximizing the potential of these sub-2-μm particle columns and design to perform under back pressures up to 15,000 psi means that separations are now routinely performed with much greater speed, resolution, and sensitivity (4). Resolution is a key factor in developing a screening method in complex samples. Food samples typically contain hundreds of components and sufficient separation of residues of interest from the matrix requires sufficient peak capacity during the sample analysis. While speed is important, it is meaningful only when sufficient separation resolution is obtained. For these reasons, ultrahigh-pressure liquid chromatography (UHPLC) is the preferred chromatographic technique for screening work.
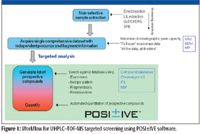
Figure 1
UHPLC coupled with quadrupole TOF-MS provides both high chromatographic peak capacity and comprehensive mass spectral datasets containing full spectrum, exact mass information. The work described here employs a quadrupole TOF-MS system that acquires spectra using a unique acquisition mode.
Using MSE to Acquire Comprehensive Datasets
The second step of our workflow (Figure 1) is to generate a comprehensive dataset with independent precursor (molecular ion) and product (fragment) ion information. This is achieved using the quadrupole TOF-MS system in MSE acquisition mode. This data acquisition strategy allows the mass spectrometer to obtain two distinct but parallel data sets, one at low collision energy (CE) and the other at high CE. The resulting raw data file contains two distinct chromatograms; one contains data with the molecular ions while the other contains fragment ion information. This means that from a single UHPLC injection, the MS full scan, containing molecular ion data, and fragment ion information can be obtained simultaneously.
Simultaneous acquisition of molecular ion and product ions improves the efficiency of the screening experiment because it enables fragment ions to be used during the targeting and identification of known compounds. MSE also provides critical structural information that assists in the identification of unknown compounds.
Processing Data with Software
After the data have been acquired, data processing is then performed using POSI±IVE software (Waters Corporation, Milford, Massachsuetts). There are two basic steps in targeted residue screening (see Figure 1). The first is simply "presence or absence screening," that is, are the compounds of interest present? If compounds are present then the second step is "quantitation": at what level are the detected compounds present in the sample? To perform the presence–absence step, the acquired dataset is interrogated using a database containing target compound information. The database, which is edited and updated easily by the user, contains target compound names and associated parameters such as the exact mass of the targeted molecular ion, exact masses of product ions, and retention time. For each compound in the database, the software will construct a narrow mass range extracted ion chromatogram (see Figure 2) within target retention time window. It also will look for the presence of specified fragment ions within the same time window. For any molecular ion peak detected in the extracted ion chromatogram, the isotopic profile will be compared with the theoretical profile based upon the compound elemental composition and the result reported as an isotope fit or "i-Fit" value. To develop effective screens (no false negatives, limited false positives) exact mass, isotopic patterns, product ions, and retention time filters are used. These are setup in the processing software and should be used to optimize method performance.
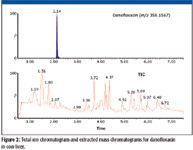
Figure 2
The result of this presence–absence screen is a list of prospective positive compounds based upon the parameters described above. It also is possible to have the software assign compounds as "tentative" detections based upon how well the retention time, mass accuracy, fragment ions, and i-Fit conditions are met. The positive (and tentative, if desired) compounds are then automatically processed using the quantification part of the software workflow.
A broad scope TOF-MS screen performed in this way requires minimal information in advance of data collection and allows the use of limited standards. This reduces labor costs by reducing handling of chemicals and time spent preparing and running extensive calibration standards. For some situations, calibration is a single point standard containing the most commonly detected target compounds at a concentration of regulatory relevance, in which case, the quantification can be regarded as semi quantitative. In this case, positive detections can then be quantified fully using a validated confirmatory method (such as MS-MS with a tandem quadrupole mass spectrometer using complete calibration curves). Alternatively, it also is feasible to run full calibration curves on the TOF-MS instrument and quantify directly.
Veterinary Drugs in Products of Animal Origin
Residual levels of veterinary drugs, widely used to treat or prevent disease in animals, are sometimes present at trace levels in meat, fish, milk, eggs, or honey. The presence of drug residues in the food chain is of concern due to their potential allergenic effects or of the potential to cause bacterial resistance to similar antibiotics in humans. To protect consumer health and to ensure the high quality of animal products, regulatory authorities have set maximum residue limits (MRLs) for allowable drug residues in animal products (5–7).
The application presented here describes the use of an ACQUITY UPLC system (Waters Corp.) coupled to a Xevo QTof quadrupole TOF-MS system (Waters Corp.) for the screening of more than 150 veterinary drug residues and metabolites including avermectins, benzimidazoles, β-agonists, β-lactams, corticosteroids, macrolides, nitroimidazoles, quinolones, sulfonamides, tetracyclines and some other veterinary medicinal products. The data were processed using the software described earlier.
Experimental
Sample Preparation
The sample preparation method for milk was reported previously (8). For liver, blood, fish, and meat samples, a 5-g sample was mixed with 20 mL acetonitrile and 5 g of anhydrous Na2SO4. After centrifugation, 0.4 mL of DMSO was added to 4 mL of supernatant. Acetonitrile was then evaporated and samples were reconstituted to 0.8 g with water and ultracentrifuged before injection.
LC Conditions
The LC separation was performed on the system mentioned previously with an ACQUITY UPLC BEH C18 column (1.7 μm, 100 mm × 2.1 mm, Waters Corp.). The column temperature was set at 40 °C. Mobile phase A was 0.1% formic acid (aqueous), and mobile phase B was acetonitrile with 0.1% formic acid. The initial gradient was held at 95% A for 0.25 min, then stepped to 5% A at 6.00 min and held for 1 min before being stepped back to 95% A at 7.20 min then held for a 1.80 min equilibration. Total run time was 9.00 min, and the injection volume was 20 μL full loop. The flow rate was kept at 0.4 mL/min.
MS Conditions
The MS detection was performed on the MS system described earlier. The data acquisition mode was MSE. The ionization mode was positive electrospray (ESI+). The source temperature was set at 120 °C, and the desolvation temperature was set at 400 °C with desolvation gas flow set at 800 L/h. The capillary voltage was set at 2.4 kV. The cone voltage was set at 30 V. The MS collision energy was set at 6 V and the MSE collision energy ramp 25–35V. Waters IntelliStart Software (Waters Corp.) for the quadrupole TOF-MS system automated the set-up of mass calibration using sodium formate and lock mass checks.
Data acquisition and processing methods
The data were acquired and processed using Waters software. In MSE acquisition mode, data is collected in two channels all of the time; low CE for molecular ion information; and high CE for product ions.
Results and Discussion
The selectivity in complex sample matrices comes from the ability to reconstruct exact mass chromatograms with narrow mass windows and is the result of the high chromatographic resolution enabled by the LC system together with the high mass spectral resolution of the quadrupole TOF-MS system. An example of the total ion chromatogram (TIC) and corresponding exact mass chromatogram for danofloxacin in a cow liver extract is illustrated in Figure 2.
High resolution is very useful for achieving selectivity and hence, confidence in results. However, in some cases, extra information, such as product ions, is of significant benefit to unambiguously identify compounds of interest. The added confidence in results is illustrated in Figure 3 by the case of xylazine and morantel, which share the same exact mass (m/z 221.1112) and elemental composition (C12H16N2S) and have similar retention times, 2.44 and 2.52 min, respectively. Xylazine and morantel share one product ion (m/z 164.0536) but can have their identity assigned with the unique product ions (m/z 90.0366 and 150.0381, respectively). Even very high mass spectral resolution would be unable to distinguish between these compounds so additional structural information must be used to unambiguously assign an identity to the residue of interest.

Figure 3
Product ion information can be obtained by performing in-source fragmentation, MS-MS, or MSE. In-source fragmentation suffers from reduced sensitivity compared to other approaches and MS-MS is a data-dependent technique that suffers from reduced duty cycle and its use requires prior knowledge of the sample contents. MSE, as described previously, is a data-independent acquisition technique that provides a simple, unbiased, parallel route to delivering both exact mass molecular ion and product ion information on every detectable component without the need for multiple injections.
Figure 4 illustrates the differences in the spectra obtained by low CE and high CE in MSE mode. The low-CE spectrum is dominated by the molecular ion while the high-CE spectrum contains product ions that add confidence to the assignment of identity.
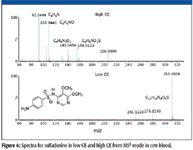
Figure 4
The samples were analyzed with a total cycle time of 9 min, including equilibration time, for a sample throughput of more than 100 samples per day. However, this level of sample throughput, together with even just 150 residues per sample, will yield significant bottlenecks in data processing.
The software used has been developed specifically to reduce the data processing burden for review of TOF-MS screening data (9) by ensuring that only positive (exact mass and retention time within definable tolerances) and tentative (retention time OK with flagging indicating exact mass out of tolerance) detections are quantified automatically. A target compound list containing compound name, formula and retention time is all that is needed. This list can be very large, enabling very many compounds to be screened.
During automated processing, the software performs a qualitative search to generate presence–absence results for the compounds in the target list, using mass accuracy and retention time to determine if compounds are positively, tentatively or negatively detected. All positive and tentative detections are then automatically quantified and listed within an application manager browser report with a measure of their isotopic fit (iFit). Figure 5 illustrates the screening results for a cow blood extract where >150 veterinary drugs have been reduced qualitatively and quantitatively to just three: hydrocortisone, sulfadoxine, and trimethoprim. The displayed sample shows the positive identification and quantification of trimethoprim in a blood extract.
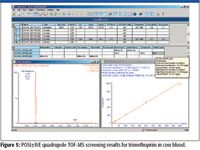
Figure 5
Figure 6 illustrates part of the report generated from a cow blood extract where 150 residues have been reduced to just three that were positively identified by retention time, exact mass, and iFIT.
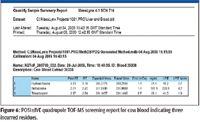
Figure 6
Conclusion
A generic workflow for residue and contaminant screening has been described using the combination of UHPLC and exact mass TOF-MS with MSE acquisition to generate a comprehensive record of precursor and fragment ion data. Dedicated software is then used to provide automated presence–absence screening and quantification of detected compounds.
The workflow was demonstrated using an LC system and a quadrupole TOF-MS system for the rapid screening of more than 150 veterinary drug residues at the relevant reporting limits in products of animal origin. MSE acquisition provides additional confidence when assigning identity to the residue of interest and overcomes the limitations of conventional data-dependent approaches.
The information-rich nature of TOF-MS data places increasing demands upon data processing software, with the reduction in manual processing and automation of repetitive tasks key to improving the quality of results and accessibility to TOF-MS.
The software significantly reduces the bottleneck of data processing for reviewing TOF-MS screening data, by ensuring that only the positive and tentative detections are quantified automatically. The automated nature of processing also reduces the possibility of errors by removing manual transcription steps from the workflow.
Tim J. Jenkins, Keith Worrall, Peter Hancock, and James Morphet are with Waters Corporation, Manchester, UK.
Emmanuelle Cognard and Didier Ortelli are with the Food Authority Control of Geneva, Geneva, Switzerland.
References
(1) "Chinese milk powder contaminated with melamine sickens 1,253 babies," The Timesonline, September 16, 2008. (http://www.timesonline.co.uk/tol/news/world/asia/article4758549.ece)
(2) "All Irish pork is recalled in dioxin poison alert," The Independent, December 7 2008. (http://www.independent.co.uk/news/world/europe/all-irish-pork-is-recalled-in-dioxin-poison-alert-1055813.html)
(3) M. Hiemstra and A. de Kok, J. Chromatogr., A 1154(1-2), 3–25 (2007).
(4) Waters White Paper, Ultra Performance LC (UPLC): New Boundaries for the Chromatography Laboratory, 720000819 EN (May 2004).
(5) http://ec.europa.eu/food/food/chemicalsafety/residues/index_en.htm
(6) http://www.foodsafety.gov/%7Efsg/animal.html
(7) http://www.mhlw.go.jp/english/topics/foodsafety/positivelist/index.html
(8) D. Ortelli, E. Cognard, P. Jan, and P. Edder, J. Chrom. B 877, 2363–2374 (2009).
(9) "POSI±IVE: Improving the Workflow for TOF Pesticide Screening" Waters Application Note 720003053en.

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
The Fundamental Role of Advanced Hyphenated Techniques in Lithium-Ion Battery Research
December 4th 2024Spectroscopy spoke with Uwe Karst, a full professor at the University of Münster in the Institute of Inorganic and Analytical Chemistry, to discuss his research on hyphenated analytical techniques in battery research.
Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.