A Study of Matrix Effects on Multiply Charged Compounds
Special Issues
In preclinical development, the absolute quantification of peptides in biological matrices becomes a challenge due to the limited availability of stable label internal standards and affinity-based cleanup. This puts a renewed emphasis on matrix effects, especially for the bioanalysis of hydrophobic peptides. While the impact of matrix effects has been studied for extensively singly charged small molecules, their effect on multiply charged compounds has yet to be characterized fully. This article discusses initial results from matrix effect experiments in relation to the bioanalysis of hydrophobic peptides and techniques used to minimize matrix effects.
In the early 1990s, high performance liquid chromatography (HPLC) coupled to mass spectrometry (MS) through the use of atmospheric pressure ionization (API) sources like electrospray ionization (ESI) revolutionized bioanalysis. LC–MS-MS replaced the traditional LC–UV technique in the bioanalytical laboratory almost overnight due to the dramatic gains in sensitivity afforded by the newer hyphenated technique. During the large-scale adoption of LC–MS-MS, a phenomenon unique to the API technique was soon discovered, in which a change in response was observed for a given concentration of a target analyte in the presence of other sample components (1). This phenomenon was termed "matrix effects." If the signal was suppressed it was called ion suppression and if the signal was enhanced, it was termed ion enhancement. The data suggests that it occurs in the early stages of the API process and many possible sources of matrix effects were identified including endogenous compounds, nonvolatile compounds such as salts, and exogenous compounds.
To overcome matrix effects, stable label internal standards were used and along with adequate sample clean-up procedures, correction factors are calculated so as to normalize response. The assessment of matrix effects is now a standard criteria and submission of matrix effects data is a requirement for the IND application submission process (3,4). Increasingly, researchers are finding that the challenge of addressing matrix effects can be addressed more efficiently by bioanalytical laboratory service providers. Strategic outsourcing to a company with experience in bioanalytical MS for example, provides access to validated assays that adrress the issue at hand. Specialist service providers will have experience in developing, validating, and utilizing assays for a range of drug development challenges.
A number of challenges must be addressed when evaluating matrix effects for peptide-based compounds. For example, with the advent of peptide-based new molecular entities, it often is not possible to get stable label internal standards early in preclinical development, and chemistry-based sample clean-up techniques might not yield the desired efficiency, renewing the emphasis on proper matrix-effect evaluation. Developing an immunoaffinity purification technique often is preferred for sample clean-up, but it is cost-prohibitive and not widely available during early development.
Hydrophobic peptides (for increased bioavailability) are of particular interest because they are eluted in a high organic portion of the mobile phase gradient, usually around the same time as endogenous plasma components such as phospholipids. Endogenous phospholipids are present to some extent in most extraction techniques used in bioanalysis and are one of the major causes of ion suppression. While the impact of endogenous phospholipids has been studied extensively for singly charged small molecules, their effect on multiply charged compounds such as peptides has yet to be characterized fully. Furthermore, while chemistry-based techniques such as solid phase extraction exploit the large difference between the chemistry of a small molecule and cellular debris such as endogenous proteins, peptides, and phospholipids, this difference in chemistries becomes smaller when trying to purify a hydrophobic peptide, limiting the extent of cleanup.
Experimental
Four peptide standards were used in this study to generate the multiply charged ions: porcine growth hormone releasing factor (GRF, FW:5108.76), glucagon-like peptide (GLP, FW: 3297.63), C-type natriuretic peptide (FW: 2197.6) (Sigma Chemicals, St. Louis, Missouri), and a proprietary peptide (FW: 1282). Intermediate solutions of 100 ng/mL each were prepared using 10 mM ammonium acetate. A 1-mg/mL solution of caffeine in methanol (Sigma Chemicals) was also used as a standard, as well as K-2 EDTA human and rat plasma (Rockland Immunochemicals, Gilbertsville, Pennsylvania).
An HPLC system (Agilent 1100 binary pump, Agilent Technologies, Santa Clara, California) was operated at a flow rate of 0.5 mL/min. Water (0.1 formic acid) was used as mobile phase A, whereas acetonitrile (0.1% formic acid) was used as mobile phase B. The gradient was set at 10% B for 1 min and it was ramped to 90% B for 3 min and held for 1 min, for a total run time of 5 min. Samples were injected on a 50 mm × 2.1 mm, 3.5-μm C4 BEH column (Waters Corp., Milford, Massachusetts) using a CTC PAL autosampler (CTC Pal, Basel, Czech Republic), with an injection volume of 20 μL.
A triple-stage quadrupole mass spectrometer (TSQ Vantage, Thermo Fisher Scientific, San Jose, California) was implemented for the quantitative analysis of the peptides using a heated electrospray ionization probe. The MS system was operated using a 4.2-kV spray voltage, 300 °C vaporizer temperature, 350 °C ion transfer tube temperature, 65 au sheath gas pressure, 15 au auxiliary gas pressure, and an 1.5 mTorr argon filled collision gas pressure. Resolution at full width half maxima (FWHM) for Q1 was set to 1.0 FWHM and Q3 was set to 0.7 FWHM using a total scan time was 0.1 s and scan width of 0.002 u.
Results and Discussion
Figure 1 shows the chromatographic elution profile for the proprietary hydrophobic peptide normalized against the elution profiles of the common endogenous phospholipids and lysophospholipids. The common phosphonate product ion (m/z 184) was monitored from precursor ions, at m/z 496, 524, 704, 758, 786, and 804 as a general indication of phospholipid elution profiles as per Meng and Bennett, Proceeedings of the ASMS Conference 2004. Figure 1 indicates the presence of a significant amount of late eluted phospholipids, which can accumulate on the column and be eluted randomly, a common issue with protein precipitation cleanup. Despite the use of C4 column chemistry, the hydrophobic peptide still is eluted in a zone close to the elution of the common phospholipids, indicating that perhaps more sample cleanup was required to develop a more robust method, if ion suppression became an issue.

Figure 1
Figure 2 shows the results of the ion suppression infusion experiment (Matuzewski et al.). While the reference small molecule (caffeine) trace clearly shows ion suppression, the trend for the multiply charged compounds (infused individually) seems to show signs of ion enhancement. While the extent of enhancement was not confirmed quantitatively, the difference in the matrix effect trend indicates a difference between singly charged small molecules and multiply charged biomolecules. One explanation based upon this initial observation could be that the multiple charges present on biomolecules create a higher electrical imbalance within the droplet and as a result, enhance ejection of gas phase ions when compared to similar droplet stoichiometry for singly charges species.
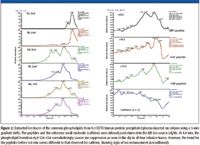
Figure 2
In the end, to ensure a robust bioanalytical method for the proprietary hydrophobic peptide, the cleanup was changed to solid-phase extraction (SPE) and ultrahigh-pressure liquid chromatography (UHPLC) was used (Figure 3). Because the method used a chemical analog internal standard, cleanup became important to ensure a high-quality bioanalytical method. The limit of quantitation was enhanced to 0.05 ng/mL using SPE and UHPLC versus 0.125 ng/mL using protein-precipitation–based sample clean-up, as shown in Figure 3.
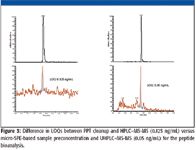
Figure 3
Conclusion
Inadequate sample clean-up, presence of nonvolatile material, and poor chromatography are contributory factors that result in matrix effects and subsequent analytical inaccuracy. Hydrophobic peptides are of particular interest as they represent an analytical challenge for traditional clean-up techniques based on extraction chemistry, such as SPE and liquid–liquid extraction, especially when working without stable label internal standards.
Rohan Thakur is with Taylor Technology, Princeton, New Jersey.
References
(1) D.L. Burhman et al., J. ASMS 7, 1099–1105 (1996).
(2) L. Jessome and D. Volmer, LCGC 24(5), 498–511 (2006).
(3) "Guidance for Industry Bioanalytical Method Validation." U.S. Department of Health and Human Services Food and Drug Administration, Center for Drug Evaluation and Research (CDER). May 2001.
(4) C.T. Viswanathan, S. Bansal, B. Booth, A.J. DeStefano, M.J. Rose, J. Sailstad, V.P. Shah, J.P. Skelly, P.G. Swann, and R. Weiner, AAPS J. 9(1), E30–42 (2007).
(5) R.C. King et al., J. ASMS 11, 942–950 (2000).

High-Speed Laser MS for Precise, Prep-Free Environmental Particle Tracking
April 21st 2025Scientists at Oak Ridge National Laboratory have demonstrated that a fast, laser-based mass spectrometry method—LA-ICP-TOF-MS—can accurately detect and identify airborne environmental particles, including toxic metal particles like ruthenium, without the need for complex sample preparation. The work offers a breakthrough in rapid, high-resolution analysis of environmental pollutants.
The Fundamental Role of Advanced Hyphenated Techniques in Lithium-Ion Battery Research
December 4th 2024Spectroscopy spoke with Uwe Karst, a full professor at the University of Münster in the Institute of Inorganic and Analytical Chemistry, to discuss his research on hyphenated analytical techniques in battery research.
Mass Spectrometry for Forensic Analysis: An Interview with Glen Jackson
November 27th 2024As part of “The Future of Forensic Analysis” content series, Spectroscopy sat down with Glen P. Jackson of West Virginia University to talk about the historical development of mass spectrometry in forensic analysis.
Detecting Cancer Biomarkers in Canines: An Interview with Landulfo Silveira Jr.
November 5th 2024Spectroscopy sat down with Landulfo Silveira Jr. of Universidade Anhembi Morumbi-UAM and Center for Innovation, Technology and Education-CITÉ (São Paulo, Brazil) to talk about his team’s latest research using Raman spectroscopy to detect biomarkers of cancer in canine sera.